Abstract
In the post genomic era, large and complex molecular datasets from genome and metagenome sequencing projects expand the limits of what is possible for bioinformatic analyses. Network-based methods are increasingly used to complement phylogenetic analysis in studies in molecular evolution, including comparative genomics, classification, and ecological studies. Using network methods, the vertical and horizontal relationships between all genes or genomes, whether they are from cellular chromosomes or mobile genetic elements, can be explored in a single expandable graph. In recent years, development of new methods for the construction and analysis of networks has helped to broaden the availability of these approaches from programmers to a diversity of users. This chapter introduces the different kinds of networks based on sequence similarity that are already available to tackle a wide range of biological questions, including sequence similarity networks, gene-sharing networks and bipartite graphs, and a guide for their construction and analyses.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
An evolutionary biologist is interested in how processes governing evolution have produced the diversity of genes, genomes, organisms, species, and communities that are observed today. For example, a biologist interested in the eukaryotes may wonder what symbiotic partners have contributed to their origins and evolution. Eukaryotic nuclear genomes are chimeric in nature, encoding many genes acquired from their alphaproteobacterial endosymbiont [1,2,3]. However, in recent years, it has been proposed that the ongoing gain of genes by both microbial [4,5,6] and multicellular eukaryotes [7, 8] via lateral gene transfer (LGT) has continued to contribute to eukaryotic evolution, though to a lesser extent than prokaryotes [9]. A biologist interested in prokaryotes may wish to investigate lateral gene transfer to explore the numbers and kinds of genes transferred between bacteria, archaea, and their mobile genetic elements [10,11,12,13,14]. These transfers are important for understanding the accessory genomes of prokaryotes [15,16,17]. Further, studying gene transfers in real bacterial communities from different environments can help to test the effect of LGT on ecology and evolution of communities [18]. Given the prevalence of introgression [9,10,11, 19], one interesting question is whether gene transfer has led to the formation of novel fusion genes that combine parts of genes originating from separate domains of life [20]. An ecologist may wish to analyze the distribution of genes and species in the environment [21]. A metagenome analyst may need to overcome an additional challenge exploring the nature of the large proportion of sequences in metagenome datasets that have little or no detectable similarity to characterize sequences and to study the “microbial dark matter” [22].
High-throughput sequencing technologies present new opportunities to investigate these diverse kinds of questions with molecular data; however, they also present challenges in terms of the scale of the analyses. Consequently, a number of network-based methods have recently been developed to expand the toolkit available to molecular biologists [23], and these have already made major contributions to our understanding of molecular evolution. Networks have been used to shed light on the nature of the “microbial dark matter” [24] and used in ecological studies to explore the geographical distribution of organisms or genes [25, 26] or the evolution of different lifestyles [27]. Their suitability for investigating introgressive events has been used to enhance our understanding of the chimeric origin of genes in the eukaryotic proteome [28, 29], the flow of genes between prokaryotes and their mobile genetic elements [30,31,32,33,34,35], and gene sharing across mobile elements to study the transfer of resistance factors [14, 36]. Networks have also been used to classify highly mosaic viral genomes [37, 38] and identify gene families [39, 40]. These approaches are highly complementary to traditional phylogenetic approaches, highlighted by the development of hybrid approaches and phylogenetic and phylogenomic networks [34, 41,42,43]. These hybrid networks are beyond the scope of discussion in this chapter but are covered in Chapters 7 and 8.
While the generation and analysis of networks were previously limited to biologists with programming experience, tools have recently been developed to simplify the process and broaden the availability of network analyses of molecular sequence data. This chapter introduces the different kinds of networks that are already available to biologists and a guide to how these networks can be constructed and analyzed for a large range of applications in molecular evolution. More precisely, this chapter will focus on three kinds of network and the types of analyses that are possible using these networks: sequence similarity networks, gene-sharing networks, and multipartite graphs [23].
2 Sequence Similarity Networks (SSNs)
Sequence similarity networks are the bread and butter of network-based molecular sequence analyses, with a huge range of applications in molecular biology. The use of SSNs for molecular sequence analysis first came to the fore in the late 1990s and early 2000s, when SSNs were suggested as a way to analyze the rapid influx of new molecular sequence data due to advances in sequencing technology and reduced cost, as well as to predict gene functions and protein-protein interactions [39, 44,45,46]. One of the earliest formal and heuristic uses of SSNs was to define the COG groups of homologous families and facilitate prediction of the functions of large numbers of genes based on homology [39, 40]. The need for efficient computation and analyses for large biological databases still pervades; however, more recently SSNs have been increasingly appreciated as useful approaches to describe complex biological systems, including inferring the “social networks” of biological life forms [30], producing maps of genetic diversity [27], detecting distant homologues [47,48,49], and exploring gene and genome rearrangements [50, 51].
A SSN is a graph in which each node is a sequence and edges connect any two nodes that are similar at the sequence level above a certain threshold (e.g., coverage, percent identity, and E-value) as determined by their pairwise alignment (Box 1) (Fig. 1). While the principle behind SSN construction is simple, the expression of similarity data in this structure can enable the use of powerful algorithms for graph analyses to study complex biological phenomena. Construction of a SSN is also frequently the starting point in a diversity of further graph analyses. A SSN can be constructed directly from fasta formatted sequence files using pipelines, such as EGN [52], the updated and faster performing EGN2 (forthcoming), or PANADA [53]. Visualization of networks can be performed with programs such as Cytoscape [54] or Gephi [55], both of which also have a range of internal tools and external plug-ins for network analysis. While these programs are useful for the visualization and analysis of relatively small networks, it can be difficult to load large and complex networks with a lot of edges (e.g., ≥50,000 edges). In these cases the iGraph library offers an extremely powerful and well-supported implementation of a broad range of commonly used methods for both complex graph generation and analysis in R, Python, and C++ [56]. However, using iGraph requires knowledge of programming in at least one of these languages. An additional package for network analysis in Python is NetworkX [57]. It is our goal here to further generalize network approaches by explaining how evolutionary biologists with less programming knowledge could analyze their data. A list including many of the tools and programs available for SSN generation is available at https://omictools.com.
Constructing a simple sequence similarity network. A set of sequences (protein or DNA) in fasta format (a) are aligned in pairs using alignment tools (such as BLAST). These alignments (b) are scored with metrics such as the percentage identity between two sequences (the number of identical nucleotides/amino acids displayed above) or the E-value of the alignment. In the resulting network (c), sequences are represented as nodes. Two sequence nodes are joined with an edge if they can be aligned above a define threshold, with the weight of the edge often based on percentage identity or E-value
Box 1: How to Build Your Own Sequence Similarity Network
-
1.
Dataset assembly: The first and most important step of SSN construction is the assembly of a dataset of sequences relevant to your biological question, usually in fasta format. This can be used as the initial input for wizards such as EGN or EGN2 [52], which can fully automate the process. The nature of the dataset is highly dependent on the research question, so here we focus on the practicalities of database assembly. To construct the similarity network, all sequences in the dataset are aligned against one another in a similarity search. This similarity search is often the time-limiting step in an analysis, and the total number of searches required is quadratic to the number of sequences in the dataset. For large datasets, it is useful to benchmark the alignment using a subset of the data to estimate the timescale for the alignment. Large datasets can generate huge outputs, not only due to the number of sequences but also the length of their identifier. One way to reduce the output size is to replace each sequence name in the fasta file with a unique integer. The use of integers will reduce disk space use and the memory consumption for any software used to analyze the sequence data.
-
2.
Similarity search: To generate a sequence similarity network, all sequences must be aligned against one another in an all-versus-all search, in which the dataset of sequences is searched against a database including the same sequences. For gene networks, the alignment is usually done with a fast pairwise aligner such as BLAST [58, 59] as implemented in EGN [52]. Filters are often used to remove low-complexity sequences from the search, as these can cause artefactual hits (BLAST options --seg yes, -soft-masking true). The BLAST method of alignment will be the focus of future discussion in this chapter; however, alternatives are available including BLAT [60] (also implemented in EGN), SWORD [61], USEARCH [62], and DIAMOND [63]. These alternatives generally include an option to produce a “BLAST” style tabulated output, making them compatible with programs commonly used in network analyses.
Within alignment tools like BLAST, it is possible to assign thresholds, such as the maximum E-value of the alignment. It is not recommended to set minimal thresholds for some parameters (such as % sequence identity) unless required due to memory constraints so that you can generate networks from a single sequence alignment with different thresholds for comparison (e.g., comparison of a 30% similarity threshold to a 90% threshold, where edges will only be drawn between highly similar genes).
Note: It may be intuitive to use additional CPUs to speed up the alignment process; however, in BLAST it can be more efficient to split the query file and launch multiple searches on separate cores instead of using the BLAST multithreading option. The pairwise alignment step is generally the most time-limiting part of generating a SSN, so benchmarking should be used to establish the optimal settings for the pairwise and/or determine the feasibility of a project given the size of the dataset and the available computational resources.
-
3.
Filtering similarity search results: In an all-versus-all similarity search, any given query sequence will have a self-hit in the corresponding database. For example, with sequences A and B, a self-hit is query sequence A matching to sequence A in the database, cases of which must be removed prior to network construction (Fig. 2). When query sequence A in a similarity search is aligned with sequence B in the database, often the reciprocal result is also identified (an alignment between query sequence B and sequence A in the database). These are called reciprocal hits; while the sequences involved are identical, the alignments and scores are not. Retaining both hits would generate two different edges between the same two nodes in a SSN, so generally only the best results from reciprocal hits are retained, based on a score such as the E-value (Fig. 2). Finally, a single query sequence may be significantly aligned multiple times in different positions of the same sequence in the database; however, for SSN construction only the best BLAST hit is generally retained (Fig. 2). The selection of the best BLAST hit is again generally often based on the E-value. Removing multiple hits against the same sequence allows the generation of an undirected network where a single edge connects two nodes, representing the best possible alignment between these nodes.
-
4.
Thresholding and network construction: Constructing a SSN from a BLAST output is conceptually simple; an edge is created between two sequences (nodes) that have been aligned in the sequence similarity search. It is common to apply thresholding criteria such as minimal % ID and/or coverage and/or maximal E-value to determine whether an edge is drawn between two sequences in the network (Fig. 1). There are different ways to calculate the % coverage of an alignment. This could be based on the coverage of a single sequence in the alignment, selecting either the query or the database sequence in each alignment or the longest or shortest sequence in each alignment. Alternatively both (mutual coverage) can be used, retaining an alignment when both values are above a given threshold. Edges above the thresholding criteria can be assigned a weight based on these criteria, producing a weighted sequence similarity network that retains information of the properties of the alignment between two sequences (Fig. 1). It is often useful to construct and compare several SSNs with variable stringencies defining the edges between sequences, for example, to optimize gene family detection within the SSN (discussed below).
Filtering sequence similarity results for network construction. In the output of an all-against-all sequence similarity search, there are a number of features that are often filtered out prior to network construction. Self-hits (1/ and 2/), where like sequences are paired in a sequence alignment, are not informative to network construction and are removed (highlighted by the red box surrounding the alignments). In cases where there are reciprocal hits (3/ and 4/) between two sequences, then only the alignment with the highest E-value is retained (highlighted with a green box around the retained alignment) to ensure only one edge representing the best possible alignment connects any two nodes in the network. The same is true for cases where a sequence has multiple hits against another sequence, such as when it aligns to another sequence in multiple positions (5/ and 6/)
2.1 Scalability of Sequence Similarity Network Analysis
As with other computational approaches, the scale of network analysis is limited by the available computational resources. The limiting factor in terms of the size of network it is possible to construct is predominantly governed by the pairwise alignment. All sequences in the dataset need to be aligned against one another in a pairwise manner, meaning the number of alignments is quadratic to the size of the dataset. For example, computing an all-against-all comparison of 1,000,000 sequences requires computation of 1012 alignments. BLAST [64] is the standard tool for this step, with a relatively good speed and accuracy for sequence similarity searches; however, the use of BLAST can be a bottleneck for the analysis of large datasets. This is an especially important consideration given the growth in the number of gene and genome sequences available in public databases. Several rapid alignment tools such as BLAT [60], USEARCH [62], Rapsearch [65], and Diamond [63] have been proposed to overcome this issue. For example, Diamond benchmarks suggest that it is almost as accurate as BLAST but is at least three orders of magnitude faster.
A second point to consider from the perspective of scalability is the complexity and size of the graph and the complexity of the algorithms used in their analysis. Algorithms where the number of calculations is linear to the size of the graph can generally be run on huge graphs with sufficient computational resources, for example, finding connected components using the “deep search first” algorithm. Algorithms for community detection (e.g., PageRank [66], Louvain) are also linear and particularly suited for detecting groups of closely related sequences in huge graphs (discussed in Subheading 4). In contrast, computing graph statistics such as the betweenness centrality are not linear to the size of the graph, even using the relatively efficient Brande algorithm for calculation [67], and are therefore more difficult to calculate for huge graphs. This has led to the development of toolkits specifically designed for the analysis of huge graphs (e.g., NetworKit) [68]. A recent book summarizes the challenges of the analysis of huge networks and some of the algorithms that have been developed to face these challenges [69].
2.2 Exploiting Sequence Similarity Networks for Identification of Gene Families
A gene family is usually defined as a group of sequences that are similar at the sequence level, indicative of homology and potentially of shared functions; however, there is no uniform way to define this similarity [70, 71]. One of the early contributions of SSNs in molecular sequence analysis was the construction of the COG database of homologous protein sequences [39, 40]. This study attempted to define gene families based on similarity at the sequence level using the results of sequence similarity searches. Within the results of an all-versus-all BLAST search, groups of at least three proteins encoded by different genomes that were more similar to each other than they were to other proteins found in the same genomes were defined as a likely orthologous gene family. Orthologous gene families are group of genes in different genomes that show sequence similarity, likely as a result of their shared evolutionary history.
The idea of using graphs to identify gene families is now a core part of many graph-based analyses. Members of a gene family aggregate in a sub-network in a SSN. These sub-networks are called connected components (CCs) at these defined thresholds, i.e., clusters of nodes connected by edges either directly or indirectly (via intermediate nodes) (Fig. 3). The size (number of nodes and edges in a CC) and density (the proportion of potential connections between all nodes in a CC that are actually connected by edges in the graph) of CCs will depend on the thresholds used for constructing the SSN as well as the relationships between sequences in the network. For example, for a given dataset at a given mutual coverage threshold, a threshold of 90% sequence identity will identify a large number of small connected components that only include highly similar genes, while at a threshold of 30% sequence identity, there will be fewer but larger connected components including genes with more variation in sequence similarity. Commonly used thresholds for detecting homologous gene families are an E-value ≤e−5, mutual coverage ≥80%, and a percentage of identity ≥30% [23].
Louvain community detection in a sequence similarity network. The network is assembled from the results of an all-versus-all alignment, as previously described. Edges can be weighted by E-value, percentage of identity, or bitscore. For the purpose of simplification, we consider strong or weak weights rather than actual values. (a) A giant connected component at relaxed threshold. (b) Three connected components at a more stringent threshold. (c) Three communities with Louvain clustering algorithm, taking into account edge weights
CCs are often detected in a SSN using the Depth-First Search (DFS) algorithm; however, there are also other approaches for the detection of gene families based on the idea of detecting “communities” [72]. In some cases, a CC can be further separated into communities of sequences that share more similarity to one another than to other sequences in the CC and thus are more highly linked in the SSN (Fig. 3). Communities are commonly identified by using graph clustering algorithms such as Louvain [73], MCL [74], or OMA [75]; however, different clustering algorithms will result in different outputs. The Louvain weighted method is widely used because it is simple to implement and scales very well to large graphs (Figs. 3 and 4) [73]. MCL is a strong deterministic algorithm that has been implemented, for example, in tribeMCL [74] and orthoMCL [76]. A potential drawback of MCL is that it requires user specification of the “inflation index,” a parameter which controls cluster granularity (or “tightness”). A high inflation index increases the tightness of clustering, producing a larger number of clusters that are smaller on average than those that would be obtained clustering the same dataset using a low inflation index. Selecting an appropriate inflation index is not trivial and requires optimization [74].
A number of the above approaches have been used to compile additional databases of orthology that can act as useful reference datasets. OMA is a program that uses graph-based algorithms and exact Smith-Waterman alignments to identify orthology between genes [77,78,79,80]. OMA is also available as a web browser [81] including a database of orthologues that, in 2015, included more than 2000 genomes and more than seven million proteins [75]. SILIX is a software package [82] that aims at building families of homologous sequences by using a transitive linkage algorithm, and HOGENOM [83] is a database that contains families inferred by SILIX for seven million proteins.
In addition to clustering genes into families, valuable information can be extracted from the connected components using network metrics. Highly conserved sequences tend to form CCs where most of the nodes are connected to each other by edges, while sequences from more divergent families will tend to form more sparsely interconnected CCs. This information can be easily assessed for each component using the clustering coefficient. Conserved families will have a clustering coefficient close to 1, even for stringent thresholds. Identifying such conserved families can be useful to produce multiple sequence alignments (MSA) needed for phylogenetic reconstruction, but SSNs have also been demonstrated to unravel relationships between distant homologues by linking distantly related sequences together [24, 29, 48]. In a SSN, two distant sequences A and C which do not share similarity according to BLAST can be linked together due to sequence B which shows similarity to both A and C.
The idea of distant homology has been particularly illuminating regarding chimeric organisms such as eukaryotes which carry homologous genes inherited from a bacterial ancestor and from an archaeal ancestor [29]. A common way to analyze sequence similarity networks is to identify certain “paths” of interest, for example, the shortest possible paths between two nodes. This notion describes the path between two nodes in a connected component that minimizes the sum of the edge weights. Alvarez-Ponce et al. used this approach to explore the topology of connected components in a SSN including the complete proteomes of 14 eukaryotes, 104 prokaryotes (including archaea and bacteria), 2389 viruses, and 1044 plasmids. Eight hundred and ninety-nine CCs contained sequences from all three domains, and of these 208 contained eukaryotic sequences that were not directly similar to one another but only linked to one another via a “eukaryote-archaea-bacteria-eukaryote” shortest path. These are putatively distant homologues in eukaryotes that were present in both the archaeal host of the mitochondrial endosymbiont and in the alphaproteobacterial endosymbiont, with both copies subsequently retained in eukaryotes and as such strong evidence for the chimeric origin of eukaryotes [29]. This demonstrates the utility of networks in the study of ancient evolutionary relationships including the origin of eukaryotes [28] or rooting the tree of life [84]. Simple path analysis for a network is possible using existing plug-ins within visualization tools such as Cytoscape [54] and Gephi [55].
2.3 Exploiting SSNs to Identify Signatures of “Tinkering” and Gene Fusion
When discussing identification of gene families, we have focused on networks where edges are drawn between protein sequences that show a high enough similarity across their entire length, defined by a high mutual coverage threshold (e.g., 80%). Sequence similarity can also be partial, for example, following gene remodeling or “tinkering” [85] producing new combinations of gene domains via gene fusion and fission events, or through the de novo sequence synthesis of gene extensions, adding to existing sequences. The term “Rosetta Stone sequence” was coined to define the formation of a new fusion protein in a species as the result of the fusion of two proteins that are found separate in another species, with authors originally predicting that these fusions could occur between proteins that physically interact in a common structural complex [86]. One of the earliest applications of sequence similarity searches to identify fusion proteins was an attempt to predict pairs of proteins that may physically interact in an organism based on whether they could be identified as a single “composite” fusion protein in another organism [44]. Beyond predicting protein-protein interactions, this kind of gene remodeling and recycling of existing gene parts has the potential to contribute to the expansion of functional diversity in genomes, creating new and unique combinations of domains and functions [51, 85, 87,88,89,90,91]. Similarity search-based screens have been implemented to identify composite genes and genome rearrangements in a range of prokaryotes [92,93,94], eukaryotes [87, 95,96,97], and viruses [98].
Early attempts to identify composite genes were based on the output of sequence similarity searches, but without formalizing the results of search methods into a graph structure. The first attempt to formalize the problem of identifying “composite” genes in networks was the “Neighborhood Correlation” approach, aiming to distinguish genuine multi-domain proteins sharing common ancestry (homologues) from novel multi-domain proteins that share domains due to insertions [99]. The later development of the FusedTriplets and MosaicFinder tools attempted to unify existing graph-based methods for detection of “composite” gene detection [50]. FusedTriplets is a graph-based implementation of the traditional gene-centered method for composite gene identification, originally introduced by Enright et al. [44], with additional cross-checks on the absence of similarity between the two component genes contributing to a composite gene based on varying thresholds [50, 100]. MosaicFinder is a gene family-centered approach which will only identify highly conserved composite gene families that form “minimal clique separators” (Fig. 5) [50]. This graph topology implies that MosaicFinder may fail to detect divergent (e.g., ancient or fast evolving) composite gene families which will tend to form “quasi-cliques” without perfect separation. CompositeSearch [101] (available at http://www.evol-net.fr/index.php/en/downloads) is a new program designed to overcome this limitation by identifying both conserved and divergent composite gene families (Box 2).
Composite gene identification using “minimal clique separators.” (a) A multiple sequence alignment of composite genes (yellow) with two components (blue and magenta). (b) The sequence similarity network corresponding to the multiple sequence alignment. The composite genes (yellow) are a minimal clique separator for the network. Their removal (shown in c) decomposes the network to the two separate component families
Box 2: How to Identify Composite Genes Using CompositeSearch
-
1.
BLAST search and filtering: An all-versus-all BLAST search is carried out as described in Box 1. Filters can be applied on the E-value and sequence similarity but should not include a mutual query coverage threshold.
-
2.
CompositeSearch: CompositeSearch takes a filtered BLAST output and a list of genes as the initial input. Two search algorithms are implemented: “fastcomposites” detects a list of potential composite genes and “composites” additionally detects potential composite gene families and component gene families. Additional options are included to filter the network based on a number of standard metrics (e.g., E-value, sequence similarity, mutual coverage) and set the maximum overlap allowed between different components aligned on the same potential composite gene. The definition of a maximum overlap allows adjustment for the tendency of BLAST to produce overhanging alignments [100]. The output includes a node, edge, and information file including information on number of nodes, edges, and family connectivity from family detection. Two outputs are included for composite gene detection, a “composites” file with detailed information on each predicted composite gene in fasta format and a “compositesinfo” file, summarizing the data. Similarly, two files provide detailed information on composite gene families and a summary of composite gene families.
-
3.
Filtering results: By default, CompositeSearch outputs all possible composite genes in “fast” mode or composite gene families in the full mode. These are given alongside a number of different metrics designed to help to filter families for more confident predictions, including the gene family size, number of composites directly predicted within the gene family, the number of domains, the number of component families, the number of singleton component families (families including only one sequence), the connectivity of the family, and a score based on the overlap between different components mapped to the composite gene.
Recent studies have explored composite gene formation as a source of innovation by “tinkering” [85] during major evolutionary transitions. These can be especially interesting when exploring genome evolution following introgression, raising the possibility of formation of new composite genes using components with different evolutionary origins [20, 51, 102]. For example, the gain of a cyanobacterial endosymbiont at the origin of photosynthetic eukaryotes was accompanied by the transfer of whole cyanobacterial genes to its new host genome, with gene functions related to the role of the plastid [103,104,105]. Identification of composite genes related to the origin of photosynthetic eukaryotes unraveled novel symbiogenetic composite genes, and unique fusions of genes encoded in the nucleus of photosynthetic eukaryotes that included components derived from the plastid endosymbiont. As with whole genes transferred to the nucleus, several of these components had predicted functions related to the role of the plastid, including redox regulations and light response [51].
2.4 Exploiting SSNs for Ecological Studies
Ecological studies increasingly involve the assembly, analysis, and comparison of large metagenome datasets. In addition to identification of functions and organisms associated with a particular environment, these studies enable the investigation of important hypotheses in microbial ecology at the level of organism or function, such as the often quoted hypothesis that “everything is everywhere, but the environment selects” from Bass Becking: the idea that microbial lineages are limitlessly dispersible in the environment, but the environmental conditions will select for certain lineages and control their distribution rather than any specific geographical separation [21].
Networks are useful for these kinds of ecological studies because existing graph algorithms can be used to investigate the structure of the network. When investigating gene (or gene-sharing networks), it is possible to distinguish nodes by labeling them based on their properties, such as categories for taxonomic or environmental origins (Fig. 6). A simple way to represent this visually is to color nodes based on these properties in Cytoscape or Gephi. A formal way to explore the relationships between node properties is to use network metrics such as conductance [106], modularity [73], and assortativity coefficient (normalized modularity) [107]. Assortativity and conductance are different metrics that attempt to answer the same type of question: do nodes labeled as belonging to a particular category, such as environmental origin, tend to be connected with other nodes labeled as belonging to the same category? More precisely, conductance quantifies whether a given category of nodes shares more edges between themselves than with nodes from different categories. A low conductance approaching zero indicates that nodes of a given category are highly connected to one another, with few connections to nodes from different categories. A higher conductance is indicative that nodes of this category tend to be more sparsely interconnected and share more connections with nodes from different categories. Assortativity is a measure of the preference for a category of nodes in a network to attach to other nodes from the same category. Normalized assortativity values range between −1 and 1, where 0 indicates random distribution of categories within the network, 1 indicates that nodes from the same categories tend to be connected to one another in the network, and −1 indicates that nodes from different categories tend to be connected in the network. A detailed description of the algorithms used in these calculations can be found in [108].
Exploring distribution of annotations in sequence similarity networks. In this example, nodes within a single connected component are assigned two colors, blue and yellow, corresponding to their having a different categorical annotation (e.g., originating from a different environmental source). Using the example of environmental source, genes in cluster A would all have the same environmental source (blue), indicating an environment-specific cluster of genes. Genes in cluster B are found in two different environmental sources (blue and yellow); however, nodes of the same type are preferentially linked to each other in the network than to genes from different environmental sources. This would result in a positive assortativity coefficient approaching 1 for environment and a low conductance score, suggesting a strong environmental community structure. Genes in cluster C are also found in two different environmental sources; however, there is no clear pattern for the distribution of genes with regard to environment. This network would have an assortativity approaching 0 and a high conductance score
2.4.1 Assortativity as a Tool to Study Geographical and Habitat Distributions of Microbes and Genes
Forster et al. used assortativity (among other network statistics, including the previously discussed shortest path analysis) to explore the geographical dispersion patterns of marine ciliates in a network generated from ciliate SSU-rDNA sequences [25]. Sequences were clustered into two different levels of gene family—CCs and Louvain communities (LCs) as previously described. Sequences were assigned categorical labels based on their geographical point of origin (eight locations) or habitat of origin (three habitats), and assortativity was calculated. If sequences, and thus species, are broadly distributed across geographical categories, then assortativity of SSU-rDNA sequences labeled with these geographical categories would be low because similar sequences would be found in different environments. Contrarily, if similar sequences tend to be from the same geographical category, indicative of endemism, then assortativity of sequence geographical origin will be high (Fig. 6). The majority of CCs and LCs showed a positive assortativity for geographical origin, higher than expected by chance, indicative of geographical community structure as opposed to global dispersal of ciliates. Similar approaches were used by Fondi et al. and applied to a collection of environmental metagenome samples to test the “everything is everywhere” hypothesis at the gene pool and functional level. Gene pools were more strongly associated with a particular ecological niche than with specific geographical location, supporting the idea that microbial genes are found everywhere but the environment selects for them [26].
2.4.2 Conductance in the Comparison of Lifestyles and Evolutionary Histories
Conductance is used to explore the clustering of pairs of different node categories in a connected component. In a study by Cheng et al., the proteomes of 84 prokaryote genomes were categorized into four broad redox groups based on their lifestyle, methanogens, obligate anaerobes, facultative anaerobes, and obligate aerobes [27]. For each CC in a pan-proteome sequence similarity network including all 84 genomes, the conductance was calculated for pairs of redox categories and compared to values obtained following random relabelling of the components. The distributions of conductance values for methanogens and for obligate anaerobes groups indicated that the sequences in these groups have features distinct from those in other groups, that anaerobes and aerobes tend to be dissimilar, and that their sequences are more isolated from one another in the SSN than expected by chance.
An additional example of the use of conductance is in exploring the propensity of a gene family to lateral gene transfer. Within a network of archaeal and bacterial genes, CCs showing a low conductance for both archaeal and bacterial sequences indicate that the bacterial and archaeal genes within the corresponding families are structured in two separate and conserved groups (Fig. 6). Structuring gene families into two groups would indicate that there was little or no evidence for lateral gene transfer between archaea and bacteria within this particular gene family. This kind of gene family is rare, with only 86 gene families from 40,584 (0.2%) meeting this criteria [24].
2.5 SSNs in Remote Homologue Identification: Shedding Light on the Microbial Dark Matter
Up to 99% of microbial species are not cultivable and thus have not been studied in isolated culture. Analysis of high-throughput sequencing and metagenomics datasets has shed light on these uncultivable organisms, often referred to as the “microbial dark matter” [109], and in some cases enabled the reconstruction of draft genomes [110,111,112,113,114]. A considerable portion of most metagenome studies have predicted ORFs showing no detectable similarity to any known proteins, termed metaORFans [115]. These can represent 25–85% of the total ORFs identified in metagenomes [22]. Identifying distant homologues of ORFans may help to predict their functions and begin to unravel the microbial dark matter. Recent work by Lopez et al. in 2015 probed the microbial diversity of metagenome datasets from a range of environments including the human gut microbiome, identifying homologues of genes from 86 ancient gene families that are distributed across archaea and bacteria. The majority of these gene families included environmental homologues that were highly divergent from any of their cultured homologues, and many branched deeply with the phylogenetic tree of life, highlighting our limited understanding of diverse elements of the microbial world and hinting at the existence of yet unknown major divisions of life [24] (Fig. 7).
Remote homologue detection to help characterize the microbial dark matter. (a) A hypothetical highly conserved cluster of genes from genomes present in sequence databases, where the average % of identity is high (≥60%). (b) The same cluster after addition of divergent environmental sequences to the network. Environmental sequences in gray are more similar to those already identified from genome surveys (≥60% max identity) so are connected directly to the conserved gene cluster in the network. More divergent sequences in pink have <60% maximum identity to their homologues in the database. Many of these are only identified as linked to the sequences from the conserved database via intermediate gray nodes. This is the notion of “transitive homology”
2.6 Exploiting SSNs to Analyze Classifications
Metagenomic and genomic data are providing scientists with a tantalizing amount of sequence data, casting the analysis of the extent of biodiversity as a major research theme in biology [116,117,118,119,120]. In theory, existing organismal and viral classifications are invaluable tools to structure and analyze this biodiversity. However, the way taxonomical classifications are constructed raises questions about their naturalness and their actual application scope [38, 120,121,122,123,124,125,126,127,128], in particular regarding genetic diversity surveys. There are three major reasons for this. First, organismal and viral diversity is still largely undersampled, which means that existing classifications are incomplete [119, 120]. Therefore, taxonomically unassigned sequences cannot be readily used in class-based genetic diversity surveys, since this dark matter remains outside existing classes. Second, classifications are constructed using different features (i.e., for viruses, a mix of phylogenetic, morphological, and structural criteria, such as replication properties in cell culture, virion morphology, serology, nucleic acid sequence, host range, pathogenicity, epidemiology, or epizootiology); therefore their classes do not necessarily offer immediate proxies for quantifying genetic diversity per se. Third, evolutionary processes responsible for both genetic and organismal diversity are diverse, and they operate at different tempos and modes in different lineages [49, 123, 129,130,131,132,133,134,135,136,137,138,139,140,141]. As a result, genetic diversity within classes and between classes can be heterogeneous, meaning that existing classifications may lack efficiency to discriminate, predict, or compare taxa on genetic bases, potentially hampering diversity studies, a profound practical issue at a time where the analysis of metagenomic sequences is becoming a priority in biology.
Addressing these challenges is notably crucial for viral studies. Recently, the executive committee of the ICTV [142] proposed that network analyses methods that create similarity metrics based on the detection of homologous genes and their genetic divergence constitute a valuable strategy to assist classification of viruses. Consistently, basic network properties and metrics (Table 1) can quantify (1) whether genetic diversity is consistent within and between the classes of existing classifications and (2) describe what classes are the most homogeneous and distinctive in terms of genetic diversity. Three criteria can be used to estimate intra-class genetic heterogeneity (Fig. 8a–c). First, the average edge weights (measured as % of identity, PID) between pairs of sequences from genomes of the same class provide a trivial measure of intra-class genetic diversity. Second, the average proportion of Conserved Canonical Connections between sequences from the same connected component and from the same taxonomic class can be exploited (CCC, i.e., in each connected component of the SSN, the total number of edges connecting sequences of a given class i (intra-group edges, denoted Eii) divided by the theoretical maximal number of possible edges between sequences of that class in the connected component (CCC(i) = 2*Eii/(N i × (N i − 1)) where N i is the number of sequences of class i present in the connected component). CCC ranges between 0 and 1. Within a connected component, if all pairs of sequences from the same class are directly connected, CCC equals 1, since all these sequences are more conserved than a given %ID threshold. By contrast, low CCC are observed when sequences from genomes from the same class lack cohesive evolution, for example, when some related sequences evolved so fast that they show less than the minimal similarity required to be directly connected to their homologues in the graph. Third, the genetic consistency of a class can be estimated by (1) identifying what cluster of sequences was present in the largest number of genomes of the class and then (2) by quantifying the proportion (in %) of the class members harboring that most ubiquitous cluster (maxCore%). When maxCore% of a class is <100%, it means that, for this dataset, there is no gene family shared by all members of that class (i.e., no core genes). The SSN structure can also serve to estimate the genetic distinctiveness of each class, i.e., whether sequences from a given class are more similar to one another than they are to sequences from other classes (Fig. 8d, e). Such sequences could be used as classificatory features to assign members to the class. In a SSN, this property translates to a low ratio of interclass edges over intra-class edges and is measured by conductance (Fig. 8d). Likewise, the proportion of clusters comprised exclusively of sequences from one class, a diagnostic feature of the class, provides an estimate of the class genetic distinctiveness. Genetically highly distinct classes have a high % of such exclusive clusters. Based on these network measures, interclass genetic heterogeneity can simply be diagnosed by contrasting estimates of genetic consistency for all the above measures for each class. There is interclass heterogeneity within a classification when the mean PID, mean CCC, maxCore%, DRC, and % of exclusive components differ between classes.
Intra- and interclasses heterogeneity measurements in weighted similarity networks. Sequences are represented by nodes. Each node is colored to represent the taxonomic class to which its host belongs. Nodes with the same color belong to the same class. Edge weight is represented by edge size proportional to the weight. Subgraphs correspond to clusters of sequences. Direct neighbors have a greater similarity than the threshold set to allow such connections. PID, average edge weights (% identity) between two sequences from genomes of the same class; CCC, average proportion of genetic conservation between sequences from the same cluster and from the same taxonomic class; maxCore%, conductance; and %-exclusive components correspond to the estimates used to assess genetic consistency of classes
Such network analyses show that virus classifications face a pragmatic issue: overall genetic distinctiveness allows relatively safe assignments of viral sequences to existing classes; however, genetic diversity of viral taxa of similar ranks differs among the tested classifications. Therefore, virus classifications (especially ICTV classification at the family level) should be used carefully to avoid inaccurate estimates in metagenomic diversity surveys. Classes with broader genetic diversity will tend to be more easily detected in the environment than classes with reduced genetic diversity, since the former will necessarily be associated with more OTUs than the latter. Some alpha- and beta-diversity analyses of environmental data, which rely on counts and on contrasts of the abundance of taxonomic classes in different samples, will thus also be biased. A similar approach could be applied on different types of classified lineages, i.e., to identify what groups of bacteria, archaea, or eukaryotes with comparable taxonomical ranks are the most genetically heterogeneous and what ranks of their classification are the least genetically consistent.
3 Gene-Sharing Networks
Gene-sharing networks are often called “genome networks” as they are best suited for summarizing what genes are shared between different genomes, highlighting routes of gene sharing. The ability to explore gene sharing between all genomes in a network in a simple graph can have useful properties for reflecting microbial social life, inherently inclusive of gene sharing both as a consequence of vertical inheritance and lateral gene transfer (LGT). Bacteriophage and plasmid genomes are typically highly mosaic in nature due to a high level of horizontal gene transfer, making it difficult to classify their genomes [37, 143]. Lima-Mendez et al. proposed the use of gene-sharing networks as a new classification method that tackles this problem of mosaicism by classifying viruses based on their genome’s content [37]. Constructing gene-sharing networks using subsets of genes from different functional categories of genes can also be useful in exploring what kinds of genes are being shared by different genomes.
In a gene-sharing network, each genome is represented by a node, and two nodes are connected by an edge when the two corresponding genomes share homologous genes or gene families (Fig. 9). These gene families can be identified from SSNs (of as CCs of LCs) or by alternative methods. In gene-sharing networks, edges can be weighted by the number of genes or gene families shared between the genomes. In this way, gene-sharing networks enable the study of microbial social life, quantitatively displaying the gene families shared between genomes both as a result of vertical transmission and lateral gene transfer.
Translating gene networks to gene-sharing networks. (a) Gene network for three gene families. Gene nodes are colored based on their genome of origin. The background color corresponds to the gene family color in part c. (b) The gene-sharing network corresponding to the gene network in a. Edges are weighted on the number of gene families shared by the genomes. (c) Multiplex gene-sharing network corresponding to the gene network in a. Genomes are connected by multiple edges with colors corresponding to different gene families. These edges are weighted based on the number of genes shared between two genomes for each family
Gene-sharing networks are useful tools for exploring overall patterns of gene sharing between genomes. Recently, Lord et al. developed BRIDES, a software package that specifically identifies different kinds of patterns in evolving gene-sharing networks after the addition of new genome nodes [144]. However, in gene-sharing networks the kind of gene families that are being shared is often overlooked. To explore how functions are shared between different genomes, gene-sharing networks can be built from genes using different subsets of functions (Fig. 10) [29]. An alternative form of the gene-sharing network is the multiplex network. In this network nodes can be linked by edges of different types, for example, each edge representing a different gene family or different functional groups of gene families, thus retaining additional information compared to a simpler gene-sharing network (Fig. 9) [23]. Multiplex networks can be useful for small-scale analyses; however, with large datasets they can rapidly become difficult to interpret and analyze. Importantly, multiplex networks are unimodal projections of bipartite graphs (discussed in the Subheading 14) which can provide greater clarity and have a number of attractive properties for the analysis of larger datasets.
Functional gene-sharing network reflecting the chimeric nature of eukaryotes. These gene-sharing networks describing how genes in different functional categories are shared between bacteria (green), archaea (yellow), eukaryotes (gray), plasmids (purple), and viruses (red) from a published dataset [29]. In both cases, a giant connected component is shown alongside examples of smaller connected components (a) Gene-sharing network for COG category D: cell division control. In this network, sequences of eukaryote origin (gray) cluster with bacterial sequences, reflecting their origin in the alphaproteobacterial endosymbiont that would become the mitochondrion. (b) Gene-sharing network for COG category K: transcription machinery. In this network, eukaryote sequence (gray) cluster with archaeal sequences, reflecting the origin of these genes in the archaeal host for the eukaryotic endosymbiont
3.1 Classification of Entities Using Gene-Sharing Networks
The possibility of summarizing gene sharing between sets of entities with complex evolutionary histories means that gene-sharing networks can be useful for classifying organisms based on their gene content. Lima-Mendez et al. analyzed bacteriophage genomes to generate two different phage gene-sharing networks that reflect their reticulate evolutionary history [37]. In the first gene-sharing network, phage genomes (nodes) were connected by edges when they shared significant similarity at the sequence level. This gene-sharing network was clustered using the previously discussed MCL algorithm [145], identifying distinct groups of phages with sequence similarity. Following clustering, membership to a particular cluster was reassessed based on shared similarity with viruses in other clusters, reflecting their reticulate evolutionary history, allowing the generation of a matrix assigning a score describing the relative membership of any given viral genome to a particular classification group. In the second approach, Lima-Mendez et al. generated a “module”-based gene-sharing network, where edges are drawn between two phage genomes if they share a “module,” in this case defined as a group of genes with similar phylogenetic profiles, enabling the exploration of what kinds of genes are shared between different groups of phages or are “signatures” for a particular group of phage genomes [37].
3.2 Exploring Routes of Gene Sharing in Gene-Sharing Networks
Two network metrics, also useful in the analysis of gene networks, can be used to attempt to identify “hubs” of gene sharing in the context of gene-sharing networks: node “degree” and “betweenness.” Both metrics aim to determine the centrality of a node in a network. The degree of a node is simply the number of edges that it is connected to. The betweenness of a node is the frequency at which it is found in all the possible shortest paths between any two nodes in the network. Halary et al. used gene-sharing networks based on DNA sequence similarity to explore gene sharing between prokaryotes and mobile genetic elements [30]. Plasmids were identified as hubs of gene sharing within this pool of genomes, suggesting that they are key vectors for genetic exchange between cellular genomes and a potential DNA reservoir shared by genomes. Phages were more peripheral in the network and mostly linked prokaryotes from the same lineage. Thus, gene-sharing networks provided insights on the evolutionary processes that shape the gene content of prokaryote genomes.
The importance of plasmids in genetic worlds was further highlighted by exploring plasmid gene-sharing networks without inclusion of prokaryote genomes [14, 36]. Connecting 2343 plasmid genomes based on shared gene content in a single graph demonstrated that plasmids tended to cluster based on the phylogenetic class of their corresponding host prokaryote rather than habitat but that more mobile plasmids tended to be more “central” in the graph, indicating that these were hubs of gene sharing. Specifically, routes of gene sharing for gene families including antibiotic resistance markers were identified between actinobacterial plasmids and gammaproteobacterial plasmids, suggesting that Actinobacteria may act as a reservoir for antibiotic resistance genes for Gammaproteobacteria [14].
The finding that plasmids are hubs of gene sharing for prokaryote genomes was supported by analysis of gene sharing in a proteobacterial phylogenomic network including 329 proteobacterial genomes [32]. A phylogenomic network is a type of phylogenetic network that has been constructed from fully sequenced genomes. In this example the phylogenomic network is an alternative to a gene-sharing network, in which genome nodes within a phylogeny are linked by edges if they share genes [34]. This study identified extensive evidence for lateral gene transfer among Proteobacteria, with at least one LGT event inferred in 75% of all gene families. Of these putative LGTs, more were related to plasmid-related genes than phage-related genes, suggesting plasmid conjugation was a more frequent source of gene transfer [32]. Directed graphs exploring directionality of LGT events between 657 prokaryote genomes allowed the polarization of 32,028 putative LGT events finding that frequency of recent events correlates with genome sequence similarity and most LGTs occurring between donor-recipient pairs with <5% difference in GC content, suggesting that there are some barriers to lateral gene transfer between prokaryotes but that these are not insurmountable [31]. Later reconstruction of transduction events linking phage donors and recipients in a phylogenomic network demonstrated that LGT by transduction was generally highest in similar genomes and between clusters of closely related species but that this constraint was occasionally broken, resulting in LGTs over long evolutionary distances [35].
4 Bipartite Graphs
Bipartite graphs are excellent at summarizing what genes are shared between sets of genomes, and as such are ideal for comparative genomics, including for the comparison of genomes reconstructed in metagenomic analyses. The potential to extend this approach to multilevel graphs, adding additional layers of information such as the environment in ecological studies, could provide a powerful summary of gene sharing in relatively complex datasets.
A multilevel network is a network in which edges exclusively connect nodes of different types, i.e., representing different levels of biological organization. Thus, a bipartite graph is a graph with two types of nodes (top and bottom nodes), where edges exclusively connect nodes of different types (Fig. 11) [146]. The types of nodes used can vary widely depending on the biological question, from linking diseases (top nodes) to their associated genes (bottom nodes) in order to explore the association between related disease phenotypes and their genetic causes [147, 148], to exploring the concept of flavor pairings in food based on a graph of ingredients (top nodes) and the flavor compounds they contain (bottom nodes) [149]. For applications in molecular biology, a typical example of a bipartite graph may describe the relationships between genomes (top nodes) and gene families (bottom nodes), with edges between nodes indicating that a genome encodes at least one member of the corresponding gene family (Fig. 11) [23, 33, 38, 150]. This kind of genome to gene family graph is particularly suited for the comparative analysis of the gene content of genomes in microbial communities and for exploring patterns of gene sharing, for example, between distantly related cellular genomes [33] or between cellular genomes and their mobile genetic elements (Corel et al. forthcoming). It is possible to represent all genes shared between a given set of genomes, as a result of both vertical inheritance and horizontal gene transfer, in a single bipartite graph [23]. This feature was utilized by Iranzo et al. to explore gene sharing among the entire dsDNA virosphere, a group of entities typified by high rates of molecular evolution and gene transfer [38]. In this case, bipartite modularity was identified in the graph to identify groups of related viral genomes and their shared genes, with the modularity of the graph optimized to Barber’s bipartite modularity [151]. A number of additional methods have been developed for detection of module structures within a bipartite graph including for weighted graphs [152]. Two recently developed tools, AcCNET [150] and MultiTwin (forthcoming), have simplified the process of constructing and analyzing multilevel graphs without the need for custom programming (Boxes 3 and 4).
A bipartite graph and its reduction to a quotient graph: (a) An example of a bipartite graph displaying how five gene families are shared between three genomes. (b) A reduced form of the bipartite graph in which gene families are combined to “twin” nodes if they share identical taxonomic distributions. A single “articulation point” connects all three genomes
Box 3: Generating Gene-Sharing Networks and Bipartite Graphs
-
1.
Dataset assembly: The same rules for dataset assembly as described in SSN generation apply to assembling the dataset for bipartite and gene-sharing graphs. It is especially important to maintain an annotation file that maps gene IDs to their genome of origin.
-
2.
Definition of gene families: Gene family identification can be carried out following the construction of sequence similarity networks, as described in Subheading 2. There are a broad range of alternative approaches for construction of gene families that are beyond the scope of discussion in this chapter; however, all of these can also be applied to the generation of gene-sharing and bipartite graphs.
-
3.
Network construction: From the definition of gene families, it is possible to construct both gene-sharing networks and bipartite graphs.
-
(a)
In a gene-sharing network, two genomes are connected by an edge when they encode genes belonging to the same gene family. Generating this kind of network can be automated from BLAST or fasta sequence data using EGN [52].
-
(b)
In a bipartite graph, there are two types of node, genome nodes and gene family nodes. An edge is drawn between a genome node and a gene family node if that genome encodes a member of the gene family. AcCNET [150] and MultiTwin (forthcoming) tools both include pipelines for generating bipartite graphs from sequence data. MultiTwin can also generate a bipartite graph from two files: a tab-delimited file mapping gene identifiers to their corresponding genome identifier and a tab-delimited file mapping gene identifiers to their corresponding gene family.
-
(a)
Two topological features of bipartite graphs can be used to facilitate studies of gene sharing by an exact decomposition of the bipartite graph: twins and articulation points [23, 153]. A bipartite graph can be reduced to a quotient graph, a reduced variant of the bipartite graph where nodes from the bipartite graph have been combined based on sharing similar properties without the loss of information. For twin nodes (“twins”), this reduction is based on the combination of bottom nodes that have identical neighbors into a single “twin” supernode in the quotient graph (Fig. 11). This is a useful way of reducing the size of large graphs without losing information, but twin nodes also have useful properties for graph interpretation. The genomes supporting a twin node (its neighbors) define a club of genomes that share genes, through common ancestry and/or horizontal transfer, and the number of gene families making up the twin gives a simple description of how many gene families are shared between this club. For example, in any given dataset, any “core” set of gene families encoded by all species in the analysis will be represented by a single twin node. The gene families combined in twin supernodes can be viewed as gene families that are likely to be transmitted together [23]. An articulation point is a node that, when removed, will split the graph into two or more connected components. Within a gene family-genome bipartite graph, articulation points are expected to help to identify “public genetic goods,” gene families that are shared by distantly related entities that may confer an advantage independent of genealogy [23, 154], as well as selfish genetic elements such as transposases that also spread across multiple genomes.
Box 4: Considerations for the Construction and Analysis of Bipartite Graphs Using AcCNET and MultiTwin
The default workflow for both ACcNet and MultiTwin takes protein sequence data in fasta format as input and generates a bipartite graph alongside a number of graph summary statistics and outputs for visualization in standard tools (such as Gephi and Cytoscape) but with a number of important differences, including:
-
Graph levels: Both AcCNET and MultiTwin can generate a bipartite graph using their default workflow; however, MultiTwin can also be used to explore additional graph levels by adding additional node types (e.g., a tripartite graph). Multipartite graphs mean that gene family level annotations can be associated with additional levels of biological information. This may be particularly useful for the comparison of samples in metagenomics studies or time course experiments, allowing gene families to be associated directly with features such as environmental origin or time point.
-
Gene family identification: AcCNET uses kClust [155] to assemble gene families, a kmer-based method for rapid assembly of clusters of homologous proteins from sequence data. By default, MultiTwin identifies gene families using an all-versus-all BLAST search, followed by identification of connected components at a given threshold, as previously discussed for gene family detection from SSNs. MultiTwin can also be used in a modular way allowing for additional customization, including the use of any custom gene family input in the form of a “community file”: a tab-delimited file linking every gene/protein ID to a community identifier, with gene families defined using a clustering method of choice.
-
Edge weighting: In AcCNET the edge weight is proportional to the inverse of the phylogenetic distance between proteins in a cluster from a given genome to other proteins within the same cluster. In MultiTwin, the default edge weight is based on the number of genes present in a gene family from any given genome.
-
Graph compression: While both methods can be used to identify “twin” nodes, only MultiTwin generates a quotient graph from these twin nodes and identifies articulation points.
AcCNET is available at: https://sourceforge.net/projects/accnet
MultiTwin is available at: http://www.evol-net.fr/index.php/en/downloads
4.1 Using Bipartite Graphs to Explore Patterns of Gene Sharing Between Diverse Entities
The simplest application of a bipartite graph is the summary of all genes shared between genomes in a single parsable graph, and this feature has been used to explore gene sharing in the dsDNA virome [38], a range of Escherichia coli genomes to investigate the E. coli pangenome [150] and between a broad range of prokaryotes that include newly discovered organisms [33]. In their analysis of prokaryote genomes, Jaffe et al. used the notion of “twins” to explore patterns of gene sharing between prokaryotes, including Archaea and the recently discovered ultrasmall “Candidate Phyla Radiation” and TM6 bacteria with extremely unusual and reduced genomes. The group found evidence for lateral gene transfer between ultrasmall bacteria and other prokaryotes, consistent with the suggestion that the ultrasmall bacteria may be symbionts [33]. In their exploration of the dsDNA virome, Iranzo et al. used graph module detection, algorithms designed to identify groups of densely connected nodes in a graph, to identify sets of densely connected viral genes and genomes that included viruses with broad host ranges, as well as 14 hallmark viral genes that account for most of the gene sharing between all different viral modules [38].
5 Conclusions
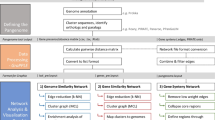
This chapter has offered a brief introduction to the generation of commonly used sequence similarity networks in molecular biology and a guide to how they can be generated and applied to a broad range of studies (Fig. 12). Networks provide a highly scalable framework for the study of an increasingly broad range of applications in molecular biology and evolution and have already contributed to a number of important discoveries in the field. These include exploring patterns of introgression and horizontal transfer across all domains of life and mobile elements, the origin of eukaryotes, the contribution of new genes including novel fusion genes to major evolutionary transitions, shedding light on the “microbial dark matter” in metagenome sequencing datasets and in testing ecological hypotheses about organism and gene distribution and environmental selection. New methods and tools for network analysis are becoming increasingly user-friendly and accessible to biologists without extensive programming experience and enabling network analysis to become a more common part of a biologist toolkit in the analysis of molecular sequence data.
6 Exercises
The exercises use EGN [52] and require access to a local installation of BLAST+ [58] and Perl. The fasta sequence file “example.faa” provided with EGN includes a dataset of protein sequences from Archaea, Bacteria, Eukaryota, and mobile genetic elements, available at http://www.evol-net.fr/index.php/fr/downloads:
-
1.
Perform a manual all-versus-all BLAST using search for a given protein sequence file from the unix terminal (requires local installation of BLAST). The output can be filtered to generate a network:
-
(a)
Make the blast database using the “makeblastdb.”
-
Command: “makeblastdb -dbtype prot -in example.faa –out example”
-
-
(b)
Performing the BLAST search using “blastp,” remembering to output data in a tabular format for easy processing.
-
Command: “blastp -query example.faa -db example -evalue 1e-5 -seg yes -soft_masking true - max_target_seqs 5000 -outfmt “6 qseqid sseqid evalue pident bitscore qstart qend qlen sstart send slen” -out protein.blastpout”
-
-
(a)
-
2.
Generate a SSN using EGN from example.faa (requires local installation of BLAST and download of EGN from http://www.evol-net.fr/index.php/fr/downloads):
-
(a)
Run EGN from the terminal using “perl egn.1.0.plus.pl” from the programs home directory.
-
(b)
Follow on-screen prompts sequentially to generate an alignment, filter the output, and generate a gene network with outputs compatible with both Cytoscape and Gephii.
-
(a)
-
3.
Visualize SSN networks:
-
(a)
In Cytoscape: Import files named “cc.*.txt” as a network to visualize that set of connected components.
-
To associate nodes with their annotations, import “cc*.atr” as a table.
-
-
(b)
In Gephi: Open “cc*.gxf” files to import individual connected components from the network into Gephi. Use the “layout” menu to explore different kinds of layouts for the network.
-
(a)
References
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135. https://doi.org/10.1038/nrg1271
Embley TM, Martin W (2006) Eukaryotic evolution, changes and challenges. Nature 440:623–630. https://doi.org/10.1038/nature04546
Williams TA, Foster PG, Cox CJ, Embley TM (2013) An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236. https://doi.org/10.1038/nature12779
Alsmark C, Foster PG, Sicheritz-Ponten T et al (2013) Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol 14:R19. https://doi.org/10.1186/gb-2013-14-2-r19
Hirt RP, Alsmark C, Embley TM (2015) Lateral gene transfers and the origins of the eukaryote proteome: a view from microbial parasites. Curr Opin Microbiol 23:155–162. https://doi.org/10.1016/j.mib.2014.11.018
Nowack ECM, Price DC, Bhattacharya D et al (2016) Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc Natl Acad Sci U S A 113:12214–12219. https://doi.org/10.1073/pnas.1608016113
McCoy JM, Mi S, Lee X et al (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. https://doi.org/10.1038/35001608
Kondo N, Nikoh N, Ijichi N et al (2002) Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci U S A 99:14280–14285. https://doi.org/10.1073/pnas.222228199
McInerney JO (2017) Horizontal gene transfer is less frequent in eukaryotes than prokaryotes but can be important (retrospective on DOI 10.1002/bies.201300095). BioEssays 39:1700002. https://doi.org/10.1002/bies.201700002
Gogarten JP, Doolittle WF, Lawrence JG (2002) Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238
Dagan T, Martin W (2007) Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci U S A 104:870–875. https://doi.org/10.1073/pnas.0606318104
Hooper SD, Mavromatis K, Kyrpides NC (2009) Microbial co-habitation and lateral gene transfer: what transposases can tell us. Genome Biol 10:R45. https://doi.org/10.1186/gb-2009-10-4-r45
Nelson-Sathi S, Sousa FL, Roettger M et al (2014) Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517:77–80. https://doi.org/10.1038/nature13805
Tamminen M, Virta M, Fani R, Fondi M (2012) Large-scale analysis of plasmid relationships through gene-sharing networks. Mol Biol Evol 29:1225–1240. https://doi.org/10.1093/molbev/msr292
Lapierre P, Gogarten JP (2009) Estimating the size of the bacterial pan-genome. Trends Genet 25:107–110. https://doi.org/10.1016/j.tig.2008.12.004
Vos M, Hesselman MC, te Beek TA et al (2015) Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol 23:598–605. https://doi.org/10.1016/j.tim.2015.07.006
McInerney JO, McNally A, O’Connell MJ (2017) Why prokaryotes have pangenomes. Nat Microbiol 2:17040. https://doi.org/10.1038/nmicrobiol.2017.40
Niehus R, Mitri S, Fletcher AG, Foster KR (2015) Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun 6:8924. https://doi.org/10.1038/ncomms9924
Hotopp JCD, Clark ME, Oliveira DCSG et al (2007) Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317:1753–1756. https://doi.org/10.1126/science.1142490
Wolf YI, Kondrashov AS, Koonin EV (2000) Interkingdom gene fusions. Genome Biol 1:research0013.1. https://doi.org/10.1186/gb-2000-1-6-research0013
Becking LB (1934) Geobiologie of inleiding tot de milieukunde. W.P. Van Stockum & Zoon, Den Haag, The Hague, the Netherlands
Lobb B, Kurtz DA, Moreno-Hagelsieb G, Doxey AC (2015) Remote homology and the functions of metagenomic dark matter. Front Genet 6:234. https://doi.org/10.3389/fgene.2015.00234
Corel E, Lopez P, Méheust R, Bapteste E (2016) Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol 24:224–237. https://doi.org/10.1016/j.tim.2015.12.003
Lopez P, Halary S, Bapteste E (2015) Highly divergent ancient gene families in metagenomic samples are compatible with additional divisions of life. Biol Direct 10:64. https://doi.org/10.1186/s13062-015-0092-3
Forster D, Bittner L, Karkar S et al (2015) Testing ecological theories with sequence similarity networks: marine ciliates exhibit similar geographic dispersal patterns as multicellular organisms. BMC Biol 13:16. https://doi.org/10.1186/s12915-015-0125-5
Fondi M, Karkman A, Tamminen MV et al (2016) “Every gene is everywhere but the environment selects”: global geolocalization of gene sharing in environmental samples through network analysis. Genome Biol Evol 8:1388–1400. https://doi.org/10.1093/gbe/evw077
Cheng S, Karkar S, Bapteste E et al (2014) Sequence similarity network reveals the imprints of major diversification events in the evolution of microbial life. Front Ecol Evol 2:72. https://doi.org/10.3389/fevo.2014.00072
Thiergart T, Landan G, Schenk M et al (2012) An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4:466–485. https://doi.org/10.1093/gbe/evs018
Alvarez-Ponce D, Lopez P, Bapteste E, McInerney JO (2013) Gene similarity networks provide tools for understanding eukaryote origins and evolution. Proc Natl Acad Sci U S A 110:E1594–E1603. https://doi.org/10.1073/pnas.1211371110
Halary S, Leigh JW, Cheaib B et al (2010) Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A 107:127–132. https://doi.org/10.1073/pnas.0908978107
Popa O, Hazkani-Covo E, Landan G et al (2011) Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res 21:599–609. https://doi.org/10.1101/gr.115592.110
Kloesges T, Popa O, Martin W, Dagan T (2011) Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol 28:1057–1074. https://doi.org/10.1093/molbev/msq297
Jaffe AL, Corel E, Pathmanathan J et al (2016) Bipartite graph analyses reveal interdomain LGT involving ultrasmall prokaryotes and their divergent, membrane-related proteins. Environ Microbiol 18:5072–5081. https://doi.org/10.1111/1462-2920.13477
Dagan T (2011) Phylogenomic networks. Trends Microbiol 19:483–491. https://doi.org/10.1016/j.tim.2011.07.001
Popa O, Landan G, Dagan T (2017) Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J 11:543–554. https://doi.org/10.1038/ismej.2016.116
Fondi M, Fani R (2010) The horizontal flow of the plasmid resistome: clues from inter-generic similarity networks. Environ Microbiol 12:3228–3242. https://doi.org/10.1111/j.1462-2920.2010.02295.x
Lima-Mendez G, Van Helden J, Toussaint A, Leplae R (2008) Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol 25:762–777. https://doi.org/10.1093/molbev/msn023
Iranzo J, Krupovic M, Koonin EV (2016) The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio 7:e00978–e00916. https://doi.org/10.1128/mBio.00978-16
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278:631–637
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. https://doi.org/10.1093/nar/28.1.33
Huson DH, Scornavacca C (2011) A survey of combinatorial methods for phylogenetic networks. Genome Biol Evol 3:23–35. https://doi.org/10.1093/gbe/evq077
Huson DH, Rupp R, Scornavacca C (2011) Phylogenetic networks: concepts, algorithms and applications. Cambridge University Press, New York, NY
Nakhleh L (2011) Evolutionary phylogenetic networks: models and issues. In: Problem solving handbook in computational biology and bioinformatics. Springer, New York, pp 125–158
Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402:86–90. https://doi.org/10.1038/47056
Pasternak G, Hochhaus A, Schultheis B, Hehlmann R (1998) Chronic myelogenous leukemia: molecular and cellular aspects. J Cancer Res Clin Oncol 124:643–660
Watanabe H, Otsuka J (1995) A comprehensive representation of extensive similarity linkage between large numbers of proteins. Bioinformatics 11:159–166. https://doi.org/10.1093/bioinformatics/11.2.159
Park J, Teichmann SA, Hubbard T, Chothia C (1997) Intermediate sequences increase the detection of homology between sequences. J Mol Biol 273:349–354. https://doi.org/10.1006/jmbi.1997.1288
Bolten E, Schliep A, Schneckener S et al (2001) Clustering protein sequences--structure prediction by transitive homology. Bioinformatics 17:935–941. https://doi.org/10.1093/bioinformatics/17.10.935
Bapteste E, Lopez P, Bouchard F et al (2012) Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc Natl Acad Sci U S A 109:18266–18272. https://doi.org/10.1073/pnas.1206541109
Jachiet P-A, Pogorelcnik R, Berry A et al (2013) MosaicFinder: identification of fused gene families in sequence similarity networks. Bioinformatics 29:837–844. https://doi.org/10.1093/bioinformatics/btt049
Méheust R, Zelzion E, Bhattacharya D et al (2016) Protein networks identify novel symbiogenetic genes resulting from plastid endosymbiosis. Proc Natl Acad Sci U S A 113:3579–3584. https://doi.org/10.1073/pnas.1517551113
Halary S, McInerney JO, Lopez P, Bapteste E (2013) EGN: a wizard for construction of gene and genome similarity networks. BMC Evol Biol 13:146. https://doi.org/10.1186/1471-2148-13-146
Martin AJM, Walsh I, Di Domenico T et al (2013) PANADA: protein association network annotation, determination and analysis. PLoS One 8:e78383. https://doi.org/10.1371/journal.pone.0078383
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Bastian M, Heymann S, Jacomy M (2009) Gephi: an Open source software for exploring and manipulating networks. Third Int AAAI Conf Weblogs Soc Media. pp 361–362. https://doi.org/10.1136/qshc.2004.010033
Csárdi G, Nepusz T (2006) The igraph software package for complex network research. InterJ Complex Syst 1695
Hagberg AA, Schult DA, Swart PJ (2008) Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux G, Vaught T, Millman J (eds) Proc. 7th Python Sci. Conf, Pasadena, CA, pp 11–15
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool.pdf. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Kent WJ (2002) BLAT--the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202. Article published online before March 2002
Vaser R, Pavlović D, Šikić M (2016) SWORD—a highly efficient protein database search. Bioinformatics 32:i680–i684. https://doi.org/10.1093/bioinformatics/btw445
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Buchfink B, Xie C, Huson DH (2014) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Ye Y, Choi J-H, Tang H (2011) RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinform 12:159. https://doi.org/10.1186/1471-2105-12-159
Page L, Brin S, Motwani R, Winograd T (1998) The PageRank citation ranking: bringing order to the web. Technical Report. Stanford InfoLab
Brandes U (2001) A faster algorithm for betweenness centrality*. J Math Sociol 25:163–177. https://doi.org/10.1080/0022250X.2001.9990249
Staudt CL, Sazonovs A, Meyerhenke H (2016) NetworKit: a tool suite for large-scale complex network analysis. Network Science 4(4):508–530. https://doi.org/10.1017/nws.2016.20
Teng S-H (2016) Scalable algorithms for data and network analysis. Now Publishers Inc, Hanover, MA
Dayhoff MO (1976) The origin and evolution of protein superfamilies. Fed Proc 35:2132–2138
Heger A, Holm L (2000) Towards a covering set of protein family profiles. Prog Biophys Mol Biol 73:321–337. https://doi.org/10.1016/S0079-6107(00)00013-4
Girvan M, Newman MEJ (2002) Community structure in social and biological networks. Proc Natl Acad Sci U S A 99:7821–7826. https://doi.org/10.1073/pnas.122653799
Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E (2008) Fast unfolding of communities in large networks. J Stat Mech Theory Exp. https://doi.org/10.1088/1742-5468/2008/10/P10008
Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. https://doi.org/10.1093/nar/30.7.1575
Altenhoff AM, Kunca N, Glover N et al (2015) The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res 43:D240–D249. https://doi.org/10.1093/nar/gku1158
Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. https://doi.org/10.1101/gr.1224503
Dessimoz C, Cannarozzi G, Gil M et al (2005) OMA, a comprehensive, automated project for the identification of orthologs from complete genome data: introduction and first achievements. Springer, Berlin, pp 61–72
Dessimoz C, Boeckmann B, Roth ACJ, Gonnet GH (2006) Detecting non-orthology in the COGs database and other approaches grouping orthologs using genome-specific best hits. Nucleic Acids Res 34:3309–3316. https://doi.org/10.1093/nar/gkl433
Roth ACJ, Gonnet GH, Dessimoz C (2008) Algorithm of OMA for large-scale orthology inference. BMC Bioinform 9:518. https://doi.org/10.1186/1471-2105-9-518
Altenhoff AM, Gil M, Gonnet GH et al (2013) Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One 8:e53786. https://doi.org/10.1371/journal.pone.0053786
Schneider A, Dessimoz C, Gonnet GH (2007) OMA browser exploring orthologous relations across 352 complete genomes. Bioinformatics 23:2180–2182. https://doi.org/10.1093/bioinformatics/btm295
Miele V, Penel S, Duret L (2011) Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinform 12:116. https://doi.org/10.1186/1471-2105-12-116
Penel S, Arigon A-M, Dufayard J-F et al (2009) Databases of homologous gene families for comparative genomics. BMC Bioinform 10:S3. https://doi.org/10.1186/1471-2105-10-S6-S3
Dagan T, Roettger M, Bryant D, Martin W (2010) Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol 2:379–392. https://doi.org/10.1093/gbe/evq025
Jacob F (1977) Evolution and tinkering. Science 196:1161–1166
Marcotte EM, Pellegrini M, Ng HL et al (1999) Detecting protein function and protein-protein interactions from genome sequences. Science 285:751–753
Kawai H, Kanegae T, Christensen S et al (2003) Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421:287–290. https://doi.org/10.1038/nature01310
Kaessmann H (2010) Origins, evolution, and phenotypic impact of new genes. Genome Res 20:1313–1326. https://doi.org/10.1101/gr.101386.109
Marsh JA, Teichmann SA (2010) How do proteins gain new domains? Genome Biol 11:126. https://doi.org/10.1186/gb-2010-11-7-126
Promponas VJ, Ouzounis CA, Iliopoulos I (2014) Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey. Brief Bioinform 15:443–454. https://doi.org/10.1093/bib/bbs072
McLysaght A, Guerzoni D (2015) New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc B Biol Sci 370:20140332. https://doi.org/10.1098/rstb.2014.0332
Enright AJ, Ouzounis CA (2000) GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics 16:451–457. https://doi.org/10.1093/bioinformatics/16.5.451
Snel B, Bork P, Huynen M (2000) Genome evolution. Gene fusion versus gene fission. Trends Genet 16:9–11
Enright AJ, Ouzounis CA (2001) Functional associations of proteins in entire genomes by means of exhaustive detection of gene fusions. Genome Biol 2:RESEARCH0034
Patthy L (2003) Modular assembly of genes and the evolution of new functions. Genetica 118:217–231
Nakamura Y, Itoh T, Martin W (2007) Rate and polarity of gene fusion and fission in Oryza sativa and Arabidopsis thaliana. Mol Biol Evol 24:110–121. https://doi.org/10.1093/molbev/msl138
Ekman D, Björklund ÅK, Elofsson A (2007) Quantification of the elevated rate of domain rearrangements in metazoa. J Mol Biol 372:1337–1348. https://doi.org/10.1016/j.jmb.2007.06.022
Jachiet P-AA, Colson P, Lopez P, Bapteste E (2014) Extensive gene remodeling in the viral world: new evidence for nongradual evolution in the mobilome network. Genome Biol Evol 6:2195–2205. https://doi.org/10.1093/gbe/evu168
Song N, Joseph JM, Davis GB et al (2008) Sequence similarity network reveals common ancestry of multidomain proteins. PLoS Comput Biol 4:e1000063. https://doi.org/10.1371/journal.pcbi.1000063
Yanai I, Derti A, DeLisi C (2001) Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.141236298
Pathmanathan JS, Lopez P, Lapointe F-J, Bapteste E (2018) CompositeSearch: a generalized network approach for composite gene families detection. Mol Biol Evol 35:252–255. https://doi.org/10.1093/molbev/msx283
Dorrell RG, Gile G, McCallum G et al (2017) Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. elife. https://doi.org/10.7554/eLife.23717
Martin W, Stoebe B, Goremykin V et al (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165. https://doi.org/10.1038/30234
Martin W, Rujan T, Richly E et al (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A 99:12246–12251. https://doi.org/10.1073/pnas.182432999
Reyes-Prieto A, Hackett JD, Soares MB et al (2006) Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr Biol. https://doi.org/10.1016/j.cub.2006.09.063
Leskovec J, Lang KJ, Dasgupta A, Mahoney MW (2008) Statistical properties of community structure in large social and information networks. In: Proceeding 17th Int. Conf. World Wide Web - WWW ’08. ACM Press, New York, p 695
Newman MEJ (2003) Mixing patterns in networks. Phys Rev E 67:26126. https://doi.org/10.1103/PhysRevE.67.026126
Newman M (2010) Networks. An introduction. Oxford University Press, Oxford. https://doi.org/10.1093/acprof:oso/9780199206650.001.0001
Rappé MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394. https://doi.org/10.1146/annurev.micro.57.030502.090759
Williams TA, Embley TM (2014) Archaeal? Dark matter? And the origin of eukaryotes. Genome Biol Evol 6:474–481. https://doi.org/10.1093/gbe/evu031
Castelle CJJ, Wrighton KCC, Thomas BCC et al (2015) Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. https://doi.org/10.1016/j.cub.2015.01.014
Brown CT, Hug LA, Thomas BC et al (2015) Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. https://doi.org/10.1038/nature14486
Spang A, Saw JH, Jørgensen SL et al (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. https://doi.org/10.1038/nature14447
Zaremba-Niedzwiedzka K, Caceres EF, Saw JH et al (2017) Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. https://doi.org/10.1038/nature21031
Prakash T, Taylor TD (2012) Functional assignment of metagenomic data: challenges and applications. Brief Bioinform 13:711–727. https://doi.org/10.1093/bib/bbs033
Hingamp P, Grimsley N, Acinas SG et al (2013) Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J 7:1678–1695. https://doi.org/10.1038/ismej.2013.59
de Vargas C, Audic S, Henry N et al (2015) Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605–1261605. https://doi.org/10.1126/science.1261605
Sunagawa S, Coelho LP, Chaffron S et al (2015) Structure and function of the global ocean microbiome. Science 348:1261359–1261359. https://doi.org/10.1126/science.1261359
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA et al (2016) Uncovering earth’s virome. Nature 536:425–430. https://doi.org/10.1038/nature19094
Shi M, Lin XD, Tian JH et al (2016) Redefining the invertebrate RNA virosphere. Nature. https://doi.org/10.1038/nature20167
van Regenmortel MH, Mayo MA, Fauquet CM, Maniloff J (2000) Virus nomenclature: consensus versus chaos. Arch Virol 145:2227–2232
Gibbs AJ (2000) Virus nomenclature descending into chaos. Arch Virol 145:1505–1507
Lawrence JG, Hatfull GF, Hendrix RW (2002) Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol 184:4891–4905
Franklin LR (2007) Bacteria, sex, and systematics. Philos Sci 74:69–95. https://doi.org/10.1086/519476
Bapteste E, Boucher Y (2008) Lateral gene transfer challenges principles of microbial systematics. Trends Microbiol 16:200–207. https://doi.org/10.1016/j.tim.2008.02.005
Bapteste E, O’Malley MA, Beiko RG et al (2009) Prokaryotic evolution and the tree of life are two different things. Biol Direct 4:34. https://doi.org/10.1186/1745-6150-4-34
Andam CP, Williams D, Gogarten JP (2010) Natural taxonomy in light of horizontal gene transfer. Biol Philos 25:589–602. https://doi.org/10.1007/s10539-010-9212-8
Koonin EV, Dolja VV (2014) Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78:278–303. https://doi.org/10.1128/MMBR.00049-13
Lederberg J, Tatum EL (1946) Gene recombination in Escherichia coli. Nature 158:558
Zinder ND, Lederberg J (1952) Genetic exchange in Salmonella. J Bacteriol 64:679–699
Levin BR (1988) Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci 319:459–472
Rodriguez-Valera F (2004) Environmental genomics, the big picture? FEMS Microbiol Lett 231:153–158
Chen I, Christie PJ, Dubnau D (2005) The ins and outs of DNA transfer in bacteria. Science 310:1456–1460. https://doi.org/10.1126/science.1114021
Edwards RA, Rohwer F (2005) Viral metagenomics. Nat Rev Microbiol 3:504–510. https://doi.org/10.1038/nrmicro1163
Frost LS, Leplae R, Summers AO, Toussaint A (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. https://doi.org/10.1038/nrmicro1235
Dagan T, Martin W (2009) Getting a better picture of microbial evolution en route to a network of genomes. Philos Trans R Soc Lond B Biol Sci 364:2187–2196. https://doi.org/10.1098/rstb.2009.0040
Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. https://doi.org/10.1146/annurev.micro.091208.073413
McDaniel LD, Young E, Delaney J et al (2010) High frequency of horizontal gene transfer in the oceans. Science 330:50. https://doi.org/10.1126/science.1192243
Dubey GP, Ben-Yehuda S (2011) Intercellular nanotubes mediate bacterial communication. Cell 144:590–600. https://doi.org/10.1016/j.cell.2011.01.015
Desnues C, La Scola B, Yutin N et al (2012) Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A 109:18078–18083. https://doi.org/10.1073/pnas.1208835109
Kutschera VE, Bidon T, Hailer F et al (2014) Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Mol Biol Evol 31:2004–2017. https://doi.org/10.1093/molbev/msu186
Simmonds P (2014) Methods for virus classification and the challenge of incorporating metagenomic sequence data. J Gen Virol. https://doi.org/10.1099/jgv.0.000016
Iranzo J, Koonin EV, Prangishvili D, Krupovic M (2016) Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsid-less mobile elements. J Virol 90:11043–11055. https://doi.org/10.1128/JVI.01622-16
Lord E, Le Cam M, Bapteste É et al (2016) BRIDES: a new fast algorithm and software for characterizing evolving similarity networks using breakthroughs, roadblocks, impasses, detours, equals and shortcuts. PLoS One 11:e0161474. https://doi.org/10.1371/journal.pone.0161474
van Dongen SM (2001) Graph clustering by flow simulation. PhD thesis, University of Utrecht
Borgatti SP, Everett MG (1997) Network analysis of 2-mode data. Soc Netw 19:243–269. https://doi.org/10.1016/S0378-8733(96)00301-2
Goh K-I, Cusick ME, Valle D et al (2007) The human disease network. Proc Natl Acad Sci U S A 104:8685–8690. https://doi.org/10.1073/pnas.0701361104
Himmelstein DS, Baranzini SE, Rand V et al (2015) Heterogeneous network edge prediction: a data integration approach to prioritize disease-associated genes. PLoS Comput Biol 11:e1004259. https://doi.org/10.1371/journal.pcbi.1004259
Ahn Y-Y, Ahnert SE, Bagrow JP et al (2011) Flavor network and the principles of food pairing. Sci Rep 1:196. https://doi.org/10.1038/srep00196
Lanza VF, Baquero F, de la Cruz F, Coque TM (2017) AcCNET (Accessory Genome Constellation Network): comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics 33:283–285. https://doi.org/10.1093/bioinformatics/btw601
Barber MJ (2007) Modularity and community detection in bipartite networks. Phys Rev E 76:66102. https://doi.org/10.1103/PhysRevE.76.066102
Beckett SJ (2016) Improved community detection in weighted bipartite networks. R Soc Open Sci 3:140536. https://doi.org/10.1098/rsos.140536
Diestel R (2010) Graph theory. Springer, New York
McInerney JO, Pisani D, Bapteste E, O’Connell MJ (2011) The public goods hypothesis for the evolution of life on Earth. Biol Direct 6:41. https://doi.org/10.1186/1745-6150-6-41
Hauser M, Mayer CE, Söding J (2013) kClust: fast and sensitive clustering of large protein sequence databases. BMC Bioinform 14:248. https://doi.org/10.1186/1471-2105-14-248
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Glossary
- Articulation point
-
A node in a graph whose removal increases the number of connected components of the resulting graph.
- Adjacency matrix
-
A numerical square matrix with row and columns labeled by network nodes, with 1 or 0 in the matrix indicating whether they are connected by an edge in the network.
- Assortativity
-
A measure of the preference for labeled nodes in a network to attach to other nodes with identical labels. This is the Pearson correlation coefficient of the degrees of pairs of linked nodes. \( \mathrm{Assortativity}=\frac{\mathrm{modularity}}{{\mathrm{modularity}}_{\mathrm{max}}} \) with modularity defined below and modularity max as the modularity of a perfectly mixed network. \( {\mathrm{modularity}}_{\mathrm{max}}=\frac{1}{2m}\left(2m-{\sum}_{ij}\frac{k_i{k}_j}{2m}\delta \left({c}_i-{c}_j\right)\right) \).
- Betweenness
-
A centrality measure for a node in a graph. Precisely, this is the proportion of shortest paths between all possible pairs of nodes in a connected component that pass through this node. A betweenness close to 1 is indicative of a highly central gene, whereas close to 0 is more peripheral.
- Bipartite graph
-
A graph with two types of nodes (top and bottom nodes), in which an edge only connects nodes of different types.
- Club of genomes
-
A group of entities that replicated separately but exploit common genetic material that may not trace back to the last common ancestor.
- Communities (also called modules)
-
In graph terminology, a community is defined as a group of nodes that are more connected between themselves than to nodes in the rest of the graph.
- Composite gene
-
A gene that is made up of at least two component parts.
- Component genes
-
Genetic fragments sharing partial similarity to a composite gene.
- Conductance
-
A measure that quantifies whether a given category of nodes shares more edges between themselves than with the rest of the nodes in the graph. A low conductance approaching zero implies that there are few edges shared between this category of nodes and the rest of the graph, while a higher conductance implies more connectivity between that category of nodes and other nodes outside of the category. G a graph, G = {V, E}. With U & G a set of nodes that is assumed to not have more than half the total node. \( \overline{U} \) = G\U. d(U) sum of degree of vertices in U. \( \mathrm{Conductance}=\frac{\sum_{i\in U,j\in \overline{U}}{a}_{i,j}}{\min \left(d(U),d\left(\overline{U}\right)\right)} \)
- Connected component
-
A subgraph in which any pair of nodes is connected, either directly or indirectly, and that is not connected to the rest of the graph.
- Degree
-
The number of edges connected to a given node.
- Endosymbiont
-
An organism that lives inside another to the mutual benefit of both organisms.
- Edge
-
The link between two nodes in a network.
- E-value
-
The number of alignments in a sequence similarity search expected to be seen by chance searching against a database of a certain size.
- Introgression
-
Descent process through which the genetic material of an entity propagates into different host structures and is replicated within these new host structures.
- Lateral gene transfer (LGT; or horizontal gene transfer, HGT)
-
Movement of genetic material between entities not mediated by vertical descent.
- Louvain community
-
A graph community identified using the Louvain algorithm. Louvain algorithm is based on optimizing modularity.
- Network (or graph)
-
A system of objects (nodes), some pairs of which are linked (edge).
- Multipartite graph
-
Similar to a bipartite graph, but with any number of types of nodes exclusively connected to nodes of other types.
- Multiplex graph
-
A graph where nodes can be connected by edges of different types.
- Modularity
-
The fraction of edges falling within given groups (e.g., communities or functional categories) in a network, minus the fraction of edges that would be expected with a random distribution of edges. With m the total number of vertices, c i the community of node i, δ() the Kronecker delta, and k i the degree of \( \mathrm{modularity}=\frac{1}{2m}{\sum}_{ij}\left({A}_{ij}-\frac{k_i{k}_j}{2m}\right)\delta \left({c}_i-{c}_j\right) \).
- Phylogenomic network
-
A phylogenetic network constructed from whole genome sequences where genomes are connected based on pairwise relationships including vertical and lateral gene transfer (LGT) events.
- Public genetic goods
-
Common genetic materials shared by clubs of phylogenetically distinct genomes.
- Quotient graph
-
A simplified graph whose nodes represent disjoint subsets of nodes of the original graph; an edge in this new graph connects two such new nodes whenever an edge in the original graph connects at least one element of a new node with at least one from the other.
- Supporting genomes
-
The common set of neighbors that support a “twin” class in a multipartite graph.
- Twins
-
Nodes in a multipartite graph that share identical sets of neighbors.
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this protocol
Cite this protocol
Watson, A.K. et al. (2019). The Methodology Behind Network Thinking: Graphs to Analyze Microbial Complexity and Evolution. In: Anisimova, M. (eds) Evolutionary Genomics. Methods in Molecular Biology, vol 1910. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9074-0_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9074-0_9
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9073-3
Online ISBN: 978-1-4939-9074-0
eBook Packages: Springer Protocols