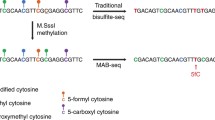
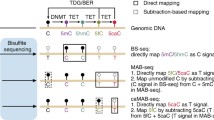
Abstract
5-Hydroxymethylcytosine (5hmC) is a modified form of cytosine, which has recently been found in mammalian cells and tissues. 5hmC is derived from 5-methylcytosine (5mC) by Ten-eleven translocation (TET) family protein-mediated oxidation and may regulate gene expression. Numerous affinity-based profiling methods have been developed to help understand the exact function of 5hmC in the genome. However, these methods have a relatively low resolution (~100 bp) without quantitative information of the modification percentage on each site. Here we demonstrated the detailed procedure of Tet-Assistant Bisulfite Sequencing (TAB-Seq), which can detect 5hmC at single-base resolution and quantify its abundance at each site. In this protocol, the genomic DNA is first treated with βGT and recombinant mTet1 consecutively to convert 5hmC to 5gmC and 5mC to 5caC, respectively. The treated genomic DNA can be directly applied to bisulfite treatment to detect 5hmC on specific loci or applied to whole-genome bisulfite sequencing as needed.
Similar content being viewed by others
References
Penn NW, Suwalski R, O'Riley C et al (1972) The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J 126:781–790
Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324:929–930
Tahiliani M, Koh KP, Shen Y et al (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935
Inoue A, Zhang Y (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334:194
He YF, Li BZ, Li Z et al (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333:1303–1307
Cortellino S, Xu J, Sannai M et al (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146:67–79
Guo JU, Su Y, Zhong C et al (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145:423–434
Globisch D, Munzel M, Muller M et al (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5:e15367
Munzel M, Globisch D, Bruckl T et al (2010) Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl 49:5375–5377
Szwagierczak A, Bultmann S, Schmidt CS et al (2010) Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 38:e181
Song CX, Szulwach KE, Fu Y et al (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29:68–72
Wu H, D’Alessio AC, Ito S et al (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25:679–684
Ito S, D’Alessio AC, Taranova OV et al (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466:1129–1133
Dawlaty MM, Ganz K, Powell BE et al (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9:166–175
Gu TP, Guo F, Yang H et al (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477:606–610
Iqbal K, Jin SG, Pfeifer GP et al (2011) Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A 108:3642–3647
Koh KP, Yabuuchi A, Rao S et al (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8:200–213
Szulwach KE, Li X, Li Y et al (2011) Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet 7:e1002154
Szulwach KE, Li X, Li Y et al (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 14:1607–1616
Kriukiene E, Liutkeviciute Z, Klimasauskas S (2012) 5-Hydroxymethylcytosine—the elusive epigenetic mark in mammalian DNA. Chem Soc Rev 41:6916–6930
Ito S, Shen L, Dai Q et al (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333:1300–1303
Pfaffeneder T, Hackner B, Truss M et al (2011) The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50:7008–7012
Maiti A, Drohat AC (2011) Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286:35334–35338
Zhang L, Lu X, Lu J et al (2012) Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8:328–330
Ficz G, Branco MR, Seisenberger S et al (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473:398–402
Williams K, Christensen J, Pedersen MT et al (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473:343–348
Xu Y, Wu F, Tan L et al (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42:451–464
Pastor WA, Pape UJ, Huang Y et al (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473:394–397
Pastor WA, Huang Y, Henderson HR et al (2012) The GLIB technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat Protoc 7:1909–1917
Huang Y, Pastor WA, Zepeda-Martinez JA et al (2012) The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat Protoc 7:1897–1908
Robertson AB, Dahl JA, Vagbo CB et al (2011) A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 39:e55
Cokus SJ, Feng S, Zhang X et al (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452:215–219
Lister R, O’Malley RC, Tonti-Filippini J et al (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536
Lister R, Pelizzola M, Dowen RH et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322
Jin SG, Kadam S, Pfeifer GP (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38:e125
Huang Y, Pastor WA, Shen Y et al (2010) The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5:e8888
Yu M, Hon GC, Szulwach KE et al (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149:1368–1380
Josse J, Kornberg A (1962) Glucosylation of deoxyribonucleic acid. III. alpha- and beta-Glucosyl transferases from T4-infected Escherichia coli. J Biol Chem 237:1968–1976
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics 27:1571–1572
Acknowledgments
This work is supported by National Institutes of Health 1R01 HG006827 to C.H.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Yu, M., Han, D., Hon, G.C., He, C. (2018). Tet-Assisted Bisulfite Sequencing (TAB-seq). In: Tost, J. (eds) DNA Methylation Protocols. Methods in Molecular Biology, vol 1708. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7481-8_33
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7481-8_33
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7479-5
Online ISBN: 978-1-4939-7481-8
eBook Packages: Springer Protocols