Abstract
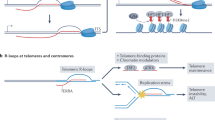
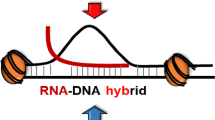
R loops are special three stranded nucleic acid structures that comprise a nascent RNA hybridized with the DNA template strand, leaving a non-template DNA single-stranded. More specifically, R loops form in vivo as G-rich RNA transcripts invade the DNA duplex and anneal to the template strand to generate an RNA:DNA hybrid, leaving the non-template, G-rich DNA strand in a largely single-stranded conformation (Aguilera and Garcia-Muse, Mol Cell 46:115–124, 2012).
DNA-RNA hybrids are a natural occurrence within eukaryotic cells, with levels of these hybrids increasing at sites with high transcriptional activity, such as during transcription initiation, repression, and elongation. RNA-DNA hybrids influence genomic instability, and growing evidence points to an important role for R loops in active gene expression regulation (Ginno et al., Mol Cell 45, 814–825, 2012; Sun et al., Science 340: 619–621, 2013; Bhatia et al., Nature 511, 362–365, 2014). Analysis of the occurrence of such structures is therefore of increasing relevance and herein we describe methods for the in vivo and in vitro identification and characterization of R loops in mammalian systems.
Abstract
R loops (DNA:RNA hybrids and the associated single-stranded DNA) have been traditionally associated with threats to genome integrity, making some regions of the genome more prone to DNA-damaging and mutagenic agents. Initially considered to be rare byproducts of transcription, over the last decade accumulating evidence has pointed to a new view in which R loops form more frequently than previously thought. The R loop field has become an increasingly expanded area of research, placing these structures as a major threat to genome stability but also as potential regulators of gene expression. Special interest has arisen as they have also been linked to a variety of diseases, including neurological disorders and cancer, positioning them as potential therapeutic targets [5].
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Aguilera A, Garcia-Muse R (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46:115–124
Ginno PA, Lott PL, Christensen HC et al (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45:814–825
Sun Q, Csorba T, Skourti-Stathaki K et al (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340:619–621
Bhatia V et al (2014) BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511:362–365
Santos-Pereira JM, Aguilera A (2015) R loops: new modulators of genome dynamics and function. Nat Rev Genet 16:583–597
Ginno PA et al (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45:814–825
Ginno PA et al (2013) GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23:1590–1600
Skourti-Stathaki K, Proudfoot NJ, Gromak N (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42:794–805
Hatchi E, Skourti-Stathaki K, Ventz S et al (2015) BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57:636–647
Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ (2014) R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516:436–439
Boque-Sastre R, Soler M, Oliveira-Mateos C et al (2015) Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A 112:5785–5790
Roy D, Lieber MR (2009) G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 29:3124–3133
Roy D et al (2010) Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol 30:146–159
Yu K, Roy D, Huang FT et al (2006) Detection and structural analysis of R-loops. Methods Enzymol 409:316–329
Yu K, Chedin F, Hsieh CL et al (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4:442–451
Acknowledgments
This work was supported by the Ministerio de Economía y Competitividad (MINECO, grant number SAF2014-56894-R), the Fundació La Marató de TV3 (grant number 20131610), and the Asociación Española contra el Cáncer-Junta de Barcelona. We are grateful to Dr. Manel Esteller for his advice and support during the preparation of this manuscript.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Boque-Sastre, R., Soler, M., Guil, S. (2017). Detection and Characterization of R Loop Structures. In: Napoli, S. (eds) Promoter Associated RNA. Methods in Molecular Biology, vol 1543. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6716-2_13
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6716-2_13
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6714-8
Online ISBN: 978-1-4939-6716-2
eBook Packages: Springer Protocols