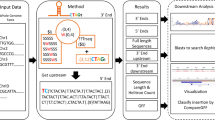
Abstract
Transposable elements (TEs) are repeat elements that can relocate or create novel copies of themselves in the genome and contribute to genomic complexity and expansion, via events such as chromosome recombination or regulation of gene expression. However, given the large number of such repeats across the genome, identifying repeats of interest can be a challenge in even well-annotated genomes, especially in more complex, TE-rich plant genomes. Here, we describe a protocol for PlanTEnrichment, a database we created comprising information on 11 plant genomes to analyze stress-associated TEs using publicly available data. By selecting a genome and providing a list of genes or genomic regions whose TE associations the user wants to identify, the user can rapidly obtain TE subfamilies found near the provided regions, as well as their superfamily and class, and the enrichment values of the repeats. The results also provide the locations of individual repeat instances found, alongside the input regions or genes they are associated with, and a bar graph of the top ten most significant repeat subfamilies identified. PlanTEnrichment is freely available at http://tools.ibg.deu.edu.tr/plantenrichment/ and can be used by researchers with rudimentary or no proficiency in computational analysis of TE elements, allowing for expedience in the identification of TEs of interest and helping further our understanding of the potential contributions of TEs in plant genomes.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bourque G, Burns KH, Gehring M et al (2018) Ten things you should know about transposable elements. Genome Biol 19:199. https://doi.org/10.1186/s13059-018-1577-z
Vicient CM, Casacuberta JM (2017) Impact of transposable elements on polyploid plant genomes. Ann Bot 120:195–207. https://doi.org/10.1093/aob/mcx078
Goerner-Potvin P, Bourque G (2018) Computational tools to unmask transposable elements. Nat Rev Genet 19:688–704. https://doi.org/10.1038/s41576-018-0050-x
Deragon JM, Casacuberta JM, Panaud O (2008) Plant transposable elements. In: Wolf J-N (ed) Plant Genomes, vol 4, pp. 69–82
Deneweth J, Van de Peer Y, Vermeirssen V (2022) Nearby transposable elements impact plant stress gene regulatory networks: a meta-analysis in A. thaliana and S. lycopersicum. BMC Genom 23:18. https://doi.org/10.1186/s12864-021-08215-8
Ramakrishnan M, Satish L, Kalendar R et al (2021) The dynamism of transposon methylation for plant development and stress adaptation. Int J Mol Sci 22:11387. https://doi.org/10.3390/ijms222111387
Nagata H, Ono A, Tonosaki K et al (2022) Temporal changes in transcripts of miniature inverted-repeat transposable elements during rice endosperm development. Plant J 109:1035–1047. https://doi.org/10.1111/tpj.15711
Karakülah G, Arslan N, Yandım C et al (2019) TEffectR: an R package for studying the potential effects of transposable elements on gene expression with linear regression model. PeerJ 7:e8192. https://doi.org/10.7717/peerj.8192
Karakülah G (2018) RTFAdb: a database of computationally predicted associations between retrotransposons and transcription factors in the human and mouse genomes. Genomics 110:257–262. https://doi.org/10.1016/j.ygeno.2017.11.002
Karakülah G, Suner A (2017) PlanTEnrichment: A tool for enrichment analysis of transposable elements in plants. Genomics 109:336–340. https://doi.org/10.1016/j.ygeno.2017.05.008
Jia S, Morton K, Zhang C et al (2018) An exome-seq based tool for mapping and selection of candidate genes in maize deletion mutants. Genom Proteom Bioinform 16:439–450. https://doi.org/10.1016/j.gpb.2018.02.003
Jia S, Yobi A, Naldrett MJ et al (2020) Deletion of maize RDM4 suggests a role in endosperm maturation as well as vegetative and stress-responsive growth. J Exp Bot 71:5880–5895. https://doi.org/10.1093/jxb/eraa325
Liao Y, Smyth GK, Shi W (2019) The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47:e47. https://doi.org/10.1093/nar/gkz114
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Makarevitch I, Waters AJ, West PT et al (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11:e1004915. https://doi.org/10.1371/journal.pgen.1004915
Forestan C, Aiese Cigliano R, Farinati S et al (2016) Stress-induced and epigenetic-mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Sci Rep 6:30446. https://doi.org/10.1038/srep30446
Lv Y, Hu F, Zhou Y et al (2019) Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response. BMC Genom 20:864. https://doi.org/10.1186/s12864-019-6245-5
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Eskier, D., Arıbaş, A., Karakülah, G. (2023). PlanTEnrichment: A How-to Guide on Rapid Identification of Transposable Elements Associated with Regions of Interest in Select Plant Genomes. In: Garcia, S., Nualart, N. (eds) Plant Genomic and Cytogenetic Databases. Methods in Molecular Biology, vol 2703. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3389-2_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3389-2_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3388-5
Online ISBN: 978-1-0716-3389-2
eBook Packages: Springer Protocols