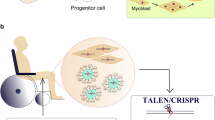
Abstract
Skeletal muscle satellite cells (SCs) are adult stem cells responsible for muscle development and injury-induced muscle regeneration. Functional elucidation of intrinsic regulatory factors governing SC activity is constrained partially by the technological limitations in editing SCs in vivo. Although the power of CRISPR/Cas9 in genome manipulation has been widely documented, its application in endogenous SCs remains largely untested. Our recent study generates a muscle-specific genome editing system leveraging the Cre-dependent Cas9 knockin mice and AAV9-mediated sgRNAs delivery, which allows gene disruption in SCs in vivo. Here, we illustrate the step-by-step procedure for achieving efficient editing using the above system.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dumont NA, Wang YX, Rudnicki MA (2015) Intrinsic and extrinsic mechanisms regulating satellite cell function. Development 142:1572–1581
Tajbakhsh S (2009) Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med 266:372–389
Goldstein JM, Tabebordbar M, Zhu K et al (2019) In situ modification of tissue stem and progenitor cell genomes. Cell Rep 27:1254–1264 e7
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Komor AC, Badran AH, Liu DR (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168:20–36
Long C, Amoasii L, Mireault AA et al (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351:400–403
Nelson CE, Hakim CH, Ousterout DG et al (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351:403–407
Nance ME, Shi R, Hakim CH et al (2019) AAV9 edits muscle stem cells in normal and dystrophic adult mice. Mol Ther 27:1568–1585
He L, Ding Y, Zhao Y et al (2021) CRISPR/Cas9/AAV9-mediated in vivo editing identifies MYC regulation of 3D genome in skeletal muscle stem cell. Stem Cell Reports 16: 2442–2458
Zhao Y, Zhou J, He L et al (2019) MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat Commun 10:5787
Guo Y, VanDusen NJ, Zhang L et al (2017) Analysis of cardiac myocyte maturation using CASAAV, a platform for rapid dissection of cardiac myocyte gene function in vivo. Circ Res 120:1874–1888
Haeussler M, Schönig K, Eckert H (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17:148
Ran FA, Hsu PD, Wright J (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Li Y, Yuan J, Chen F et al (2020) Long noncoding RNA SAM promotes myoblast proliferation through stabilizing Sugt1 and facilitating kinetochore assembly. Nat Commun 11:1–16
Zhu S, Li W, Liu J et al (2016) Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat Biotechnol 34:1279–1286
Grieger JC, Choi VW, Samulski RJ (2006) Production and characterization of adeno-associated viral vectors. Nat Protoc 1:1412–1428
Kimura T, Ferran B, Tsukahara Y et al (2019) Production of adeno-associated virus vectors for in vitro and in vivo applications. Sci Rep 9:1–13
Liu L, Cheung TH, Charville G et al (2015) Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc 10:1612–1624
Guo T, Feng YL, Xiao JJ et al (2018) Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol 19:170
Aloisio GM, Nakada Y, Saatcioglu HD et al (2014) PAX7 expression defines germline stem cells in the adult testis. J Clin Invest 124:3929–3944
Smits AH, Ziebell F, Joberty G et al (2019) Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 16:1087–1093
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Table S1
Sequences of primers used in this protocol (DOCX 85 kb)
Rights and permissions
Copyright information
© 2023 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
He, L., He, Z., Li, Y., Sun, H., Wang, H. (2023). In Vivo Investigation of Gene Function in Muscle Stem Cells by CRISPR/Cas9-Mediated Genome Editing. In: Asakura, A. (eds) Skeletal Muscle Stem Cells. Methods in Molecular Biology, vol 2640. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3036-5_21
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3036-5_21
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3035-8
Online ISBN: 978-1-0716-3036-5
eBook Packages: Springer Protocols