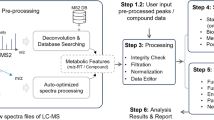
Abstract
Advances in computational and data processing technology have enabled the development of many novel tools for analyzing metabolomic and lipidomic data. These advances involved the catalyst for the creation of publicly accessible complex web-based databases such as the Metabolomics Workbench. Open Source internet-based software packages such as MetaboAnalyst 5.0 enable researchers to perform a wide range of analyte identification and statistical analyses of their own and other researchers’ data in order to identify biomarkers and classify compounds. In this paper, we set forth a protocol for obtaining experimental data of interest from a public data repository (Metabolomics Workbench), converting the data into a format suitable for submission to MetaboAnalyst 5.0, and then uploading the data to the MetaboAnalyst server for identification and statistical analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Proper S, Azouz N, Mersha T (2021) Achieving precision medicine in allergic disease: progress and challenges. Front Immunol 12:720746
Manchester M, Anand A (2017) Chapter two – metabolomics: strategies to define the role of metabolism in virus infection and pathogenesis. In: Kielian M, Mettenleiter TC, Roossinck MJ (eds) Advances in virus research. Academic Press, London, pp 57–81
Pang Z et al (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49(W1):W388–W396
Sud M et al (2016) Metabolomics workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 44(D1):D463–D470
Roberts LD et al (2012) Targeted metabolomics. Curr Protoc Mol Biol Chapter 30: p Unitas 30(2):1–24
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Howell, A., Yaros, C. (2023). Downloading and Analysis of Metabolomic and Lipidomic Data from Metabolomics Workbench Using MetaboAnalyst 5.0. In: Bhattacharya, S.K. (eds) Lipidomics. Methods in Molecular Biology, vol 2625. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2966-6_26
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2966-6_26
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2965-9
Online ISBN: 978-1-0716-2966-6
eBook Packages: Springer Protocols