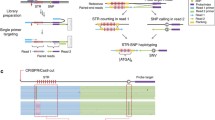
Abstract
Microsatellites are short tandem repeats of one to six nucleotides that are highly polymorphic and extensively used as genetic markers in numerous biomedical applications, including the detection of microsatellite instability (MSI) in cancer. The standard analytical method for microsatellite analysis relies on PCR amplification followed by capillary electrophoresis or, more recently, next-generation sequencing (NGS). However, their amplification during PCR generates undesirable frameshift products known as stutter peaks caused by polymerase slippage, complicating data analysis and interpretation, while very few alternative methods for microsatellite amplification have been developed to reduce the formation of these artifacts. In this context, the recently developed low-temperature recombinase polymerase amplification (LT-RPA) is an isothermal DNA amplification method at low temperature (32 °C) that drastically reduces and sometimes completely abolishes the formation of stutter peaks. LT-RPA greatly simplifies the genotyping of microsatellites and improves the detection of MSI in cancer. In this chapter, we describe in detail all the experimental steps necessary for the development of LT-RPA simplex and multiplex assays for microsatellite genotyping and MSI detection, including the design, optimization, and validation of the assays combined with capillary electrophoresis or NGS.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Change history
23 June 2023
A correction has been published.
References
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5(6):435–445. https://doi.org/10.1038/nrg1348
Gulcher J (2012) Microsatellite markers for linkage and association studies. Cold Spring Harb Protoc 2012(4):425–432. https://doi.org/10.1101/pdb.top068510
Putman AI, Carbone I (2014) Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol Evol 4(22):4399–4428. https://doi.org/10.1002/ece3.1305
Stadele V, Vigilant L (2016) Strategies for determining kinship in wild populations using genetic data. Ecol Evol 6(17):6107–6120. https://doi.org/10.1002/ece3.2346
Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Islam KN, Latif MA (2013) A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci 14(11):22499–22528. https://doi.org/10.3390/ijms141122499
Gettings KB, Aponte RA, Vallone PM, Butler JM (2015) STR allele sequence variation: current knowledge and future issues. Forensic Sci Int Genet 18:118–130. https://doi.org/10.1016/j.fsigen.2015.06.005
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138(6):2073–2087. e2073. https://doi.org/10.1053/j.gastro.2009.12.064
Mirkin SM (2007) Expandable DNA repeats and human disease. Nature 447(7147):932–940. https://doi.org/10.1038/nature05977
Hause RJ, Pritchard CC, Shendure J, Salipante SJ (2016) Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 22(11):1342–1350. https://doi.org/10.1038/nm.4191
Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S (2004) Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96(4):261–268
Wimmer K, Kratz CP, Vasen HF, Caron O, Colas C, Entz-Werle N, Gerdes AM, Goldberg Y, Ilencikova D, Muleris M, Duval A, Lavoine N, Ruiz-Ponte C, Slavc I, Burkhardt B, Brugieres L (2014) Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J Med Genet 51(6):355–365. https://doi.org/10.1136/jmedgenet-2014-102284
Zeinalian M, Hashemzadeh-Chaleshtori M, Salehi R, Emami MH (2018) Clinical aspects of microsatellite instability testing in colorectal cancer. Adv Biomed Res 7:28. https://doi.org/10.4103/abr.abr_185_16
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413. https://doi.org/10.1126/science.aan6733
Zhang S, Niu Y, Bian Y, Dong R, Liu X, Bao Y, Jin C, Zheng H, Li C (2018) Sequence investigation of 34 forensic autosomal STRs with massively parallel sequencing. Sci Rep 8(1):6810. https://doi.org/10.1038/s41598-018-24495-9
Waalkes A, Smith N, Penewit K, Hempelmann J, Konnick EQ, Hause RJ, Pritchard CC, Salipante SJ (2018) Accurate pan-cancer molecular diagnosis of microsatellite instability by single-molecule molecular inversion probe capture and high-throughput sequencing. Clin Chem 64(6):950–958. https://doi.org/10.1373/clinchem.2017.285981
Baudrin LG, Deleuze JF, How-Kit A (2018) Molecular and computational methods for the detection of microsatellite instability in cancer. Front Oncol 8:621. https://doi.org/10.3389/fonc.2018.00621
Hauge XY, Litt M (1993) A study of the origin of ‘shadow bands’ seen when typing dinucleotide repeat polymorphisms by the PCR. Hum Mol Genet 2(4):411–415
Murray V, Monchawin C, England PR (1993) The determination of the sequences present in the shadow bands of a dinucleotide repeat PCR. Nucleic Acids Res 21(10):2395–2398
Gill P, Haned H, Bleka O, Hansson O, Dorum G, Egeland T (2015) Genotyping and interpretation of STR-DNA: low-template, mixtures and database matches-twenty years of research and development. Forensic Sci Int Genet 18:100–117. https://doi.org/10.1016/j.fsigen.2015.03.014
How-Kit A, Daunay A, Buhard O, Meiller C, Sahbatou M, Collura A, Duval A, Deleuze JF (2018) Major improvement in the detection of microsatellite instability in colorectal cancer using HSP110 T17 E-ice-COLD-PCR. Hum Mutat 39(3):441–453. https://doi.org/10.1002/humu.23379
Bright JA, Taylor D, Curran JM, Buckleton JS (2013) Developing allelic and stutter peak height models for a continuous method of DNA interpretation. Forensic Sci Int Genet 7(2):296–304. https://doi.org/10.1016/j.fsigen.2012.11.013
Coble MD, Bright JA (2019) Probabilistic genotyping software: an overview. Forensic Sci Int Genet 38:219–224. https://doi.org/10.1016/j.fsigen.2018.11.009
Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC (2014) Microsatellite instability detection by next generation sequencing. Clin Chem 60(9):1192–1199. https://doi.org/10.1373/clinchem.2014.223677
Daunay A, Duval A, Baudrin LG, Buhard O, Renault V, Deleuze JF, How-Kit A (2019) Low temperature isothermal amplification of microsatellites drastically reduces stutter artifact formation and improves microsatellite instability detection in cancer. Nucleic Acids Res 47:e141. https://doi.org/10.1093/nar/gkz811
Daher RK, Stewart G, Boissinot M, Bergeron MG (2016) Recombinase polymerase amplification for diagnostic applications. Clin Chem 62(7):947–958. https://doi.org/10.1373/clinchem.2015.245829
Piepenburg O, Williams CH, Stemple DL, Armes NA (2006) DNA detection using recombination proteins. PLoS Biol 4(7):e204. https://doi.org/10.1371/journal.pbio.0040204
Hite JM, Eckert KA, Cheng KC (1996) Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n.d(G-T)n microsatellite repeats. Nucleic Acids Res 24(12):2429–2434
Dorard C, de Thonel A, Collura A, Marisa L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O, Gobbo J, Penard-Lacronique V, Zouali H, Tubacher E, Kirzin S, Selves J, Milano G, Etienne-Grimaldi MC, Bengrine-Lefevre L, Louvet C, Tournigand C, Lefevre JH, Parc Y, Tiret E, Flejou JF, Gaub MP, Garrido C, Duval A (2011) Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med 17(10):1283–1289. https://doi.org/10.1038/nm.2457
Buhard O, Lagrange A, Guilloux A, Colas C, Chouchene M, Wanherdrick K, Coulet F, Guillerm E, Dorard C, Marisa L, Bokhari A, Greene M, El-Murr N, Bodo S, Muleris M, Sourouille I, Svrcek M, Cervera P, Blanche H, Lefevre JH, Parc Y, Lepage C, Chapusot C, Bouvier AM, Gaub MP, Selves J, Garrett K, Iacopetta B, Soong R, Hamelin R, Garrido C, Lascols O, Andre T, Flejou JF, Collura A, Duval A (2016) HSP110 T17 simplifies and improves the microsatellite instability testing in patients with colorectal cancer. J Med Genet 53(6):377–384. https://doi.org/10.1136/jmedgenet-2015-103518
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3--new capabilities and interfaces. Nucleic Acids Res 40(15):e115. https://doi.org/10.1093/nar/gks596
How-Kit A, Tost J (2015) Pyrosequencing(R)-based identification of low-frequency mutations enriched through enhanced-ice-COLD-PCR. Methods Mol Biol 1315:83–101. https://doi.org/10.1007/978-1-4939-2715-9_7
Acknowledgments
We want to thank Steven McGinn (CNRGH) for his careful editing of the manuscript and improvement of the English. This work has been supported by the Fondation ARC pour la recherche sur le cancer (PJA 20191209442). LMH received support from the GENMED Laboratory of Excellence on Medical Genomics [ANR-10-LABX-0013]. SIJ and VR contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Jeanjean, S.I. et al. (2023). LT-RPA: An Isothermal DNA Amplification Approach for Improved Microsatellite Genotyping and Microsatellite Instability Detection. In: Myers, M.B., Schandl, C.A. (eds) Clinical Applications of Nucleic Acid Amplification. Methods in Molecular Biology, vol 2621. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2950-5_7
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2950-5_7
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2949-9
Online ISBN: 978-1-0716-2950-5
eBook Packages: Springer Protocols