Abstract
The mammary gland consists of a bilayered epithelial structure with an extensively branched morphology. The majority of this epithelial tree is laid down during puberty, during which actively proliferating terminal end buds repeatedly elongate and bifurcate to form the basic structure of the ductal tree. Mammary ducts consist of a basal and luminal cell layer with a multitude of identified sub-lineages within both layers. The understanding of how these different cell lineages are cooperatively driving branching morphogenesis is a problem of crossing multiple scales, as this requires information on the macroscopic branched structure of the gland, as well as data on single-cell dynamics driving the morphogenic program. Here we describe a method to combine genetic lineage tracing with whole-gland branching analysis. Quantitative data on the global organ structure can be used to derive a model for mammary gland branching morphogenesis and provide a backbone on which the dynamics of individual cell lineages can be simulated and compared to lineage-tracing approaches. Eventually, these quantitative models and experiments allow to understand the couplings between the macroscopic shape of the mammary gland and the underlying single-cell dynamics driving branching morphogenesis.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Branching morphogenesis
- Mammary gland
- Whole-mount imaging
- Clonal lineage tracing
- Biophysical modeling
- Stem cell fate choices
1 Introduction
Throughout evolution, branching has proven\ to be an efficient way to maximize the surface of exchange between the interior and exterior of an organ or organism and can therefore be found everywhere in nature [1]. Examples of this include branched tree crowns but also root systems and coral reefs. Moreover, many organs within higher organisms (including humans) are organized in treelike structures, including the lungs, kidneys, prostate, liver, pancreas, the circulatory system, and the mammary gland.
The mammary gland consists of a bilayered epithelial structure, which, originating from the nipple, forms an extensively branched tree surrounded by a fat pad and stroma. After an initial phase of branching during embryogenesis, the majority of this epithelial tree is laid down throughout puberty, during which actively proliferating terminal end buds, also referred to as tips, repeatedly elongate and bifurcate to form the basic structure of the ductal tree. After pubertal branching morphogenesis, the mammary gland undergoes continuous morphological changes throughout the life span of the organism, such as cyclic side branching driven by the estrous cycle or expansion and alveolar differentiation during pregnancy. Interestingly, mammary glands are characterized by profound structural heterogeneity, indicating that no deterministic or rigid program is driving pubertal branching morphogenesis [2, 3].
The bilayered mammary epithelium consists of a basal (also referred to as myoepithelial) and luminal cell layer but harbors many different sub-lineages within both layers, which are thought to have specific roles during different developmental stages [4]. The understanding of how these different cell lineages are cooperatively driving branching morphogenesis is a problem of crossing multiple scales, as this requires information on overall branching structure, as well as individual cellular dynamics. At the organ level, quantitative analysis of the branched morphology is required to derive a model for branching morphogenesis. The model can subsequently provide a backbone on which cellular dynamics of individual cell lineages can be simulated. Quantitative data on how different cell types or lineage contribute to branching morphogenesis can be obtained through genetic lineage-tracing approaches [3]. Eventually, information on the cellular dynamics obtained by clonal lineage tracing needs to be coupled to their relative location within the ductal tree, as well as to quantitative information on the branched morphology of the organ to understand how different cell lineages contribute to the branched morphology of the mammary gland.
The here described whole-mount analysis methods aim to understand both the emergence of macroscopic organ shape and the underlying single-cell dynamics driving the branching morphogenesis program. Although it is known that many more factors are at play, such as rigidity and differences in the structure of the mesenchyme surrounding the epithelium [5] or local signaling from immune cells and stromal cells [6], this reductionist approach allows to model how single-cell dynamics drive such a complex process and can serve as the basis for more advanced studies on branching morphogenesis. This protocol briefly describes the basic concepts of lineage tracing, after which the procedures of mammary gland whole-mount staining, imaging, analysis and model-data comparisons are described in detail.
2 Materials
2.1 Genetic Lineage Tracing
-
1.
Mouse strains carrying a Cre-inducible fluorescent reporter allele combined with a cell lineage-specific or ubiquitous Cre recombinase driver allele.
-
2.
Microbalance.
-
3.
Tamoxifen dissolved in sunflower oil or doxycycline dissolved in phosphate-buffered saline (PBS) (depending on the preferred mouse line).
-
4.
1 mL syringe with 25G × 5/8 needle (slip tip).
2.2 Mammary Gland Dissection
-
1.
Dissection pad and pins.
-
2.
70% ethanol.
-
3.
Pair of small dissection scissors.
-
4.
Two pairs of blunt forceps.
-
5.
Ice-cold PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 in 800 mL demineralized water. Adjust the pH to 7.4 and fill up with demineralized water to 1 L.
2.3 Whole-Mount Preparation
-
1.
Microbalance.
-
2.
pH meter.
-
3.
12- or 24-wells plate.
-
4.
Aluminum foil.
-
5.
Rocking plate.
-
6.
PBS.
-
7.
Collagenase A derived from Clostridium histolyticum (Sigma-Aldrich, 10103586001).
-
8.
Hyaluronidase (Sigma-Aldrich, H3506).
-
9.
4% paraformaldehyde (PFA) in 1x PBS.
-
10.
0.2 M Na2HPO4: 28.39 g/L demi H20 (store at room temperature, sterile).
-
11.
0.2 M NaH2PO4: 24 g/L demi H20 (store at room temperature, sterile).
-
12.
P-buffer: mix 81 mL of 0.2 M Na2HPO4 with 19 mL of 0.2 M NaH2PO4 and add 100 mL of demi H20. Adjust pH to 7.4 if necessary. Store at 4 °C.
-
13.
0.2 M L-lysine in P-buffer: 5.848 g in 200 mL P-buffer. Adjust pH to 7.4 if necessary. Store at 4 °C.
-
14.
Sodium periodate (NaIO4).
-
15.
PLP-buffer: To make 10 mL, mix 2.5 mL of 4% PFA with 0.0212 g of NaIO4. Shake thoroughly to dissolve the NaIO4. Next, add 3.75 mL of 0.2 M L-Lysine and 3.75 mL of P-buffer and gently shake to mix all components.
-
16.
Bovine serum albumin (BSA).
-
17.
Triton X-100.
-
18.
Blocking/permeabilization buffer: add 1% BSA, 0.8% Triton X-100, and 5% normal goat serum or 5% fetal calf serum to PBS.
-
19.
PBS with 0.2% Tween 20.
-
20.
Normal goat serum or fetal calf serum.
-
21.
Anti-fade mounting medium.
-
22.
Coverslips 60 × 24 mm, No. 1.5.
-
23.
Whole-gland mounting device (see also Note 14).
-
24.
Nail polish.
2.4 Image Visualization and Analysis
-
1.
Volumetric rendering software: LASX 3D Visualization (Leica Microsystems), Imaris (Bitplane AG), Arivis Vision4D (ZEISS), Amira (Visage Imaging), or other commercially available software packages.
-
2.
Tree surveyor [7].
-
3.
Branch analysis software [2], available upon request.
-
4.
Ete2 Python toolkit for tree visualization (etetoolkit.org)
3 Methods
3.1 Genetic Lineage Tracing to Mark Contribution of Cell Population(s) of Interest
Before starting a genetic lineage-tracing experiment, several important points need to be considered. It is important to carefully design such an experiment, as flaws in the design rapidly lead to erroneous conclusions. The following steps provide a concise guide on the design of a lineage-tracing experiment in the mammary gland, for further reading [8,9,10] are recommended.
-
1.
Choose the appropriate reporter strain. First, determine a suitable and inducible reporter mouse strain to genetically label the cell population of interest with (a) fluorescent protein(s) [11] (see Note 1).
-
2.
Choose the appropriate mode of Cre induction. Expression of Cre recombinase to activate the reporter allele at the desired time and location can be achieved in multiple ways. To label a specific cell population of interest, the reporter mouse strain can be crossed with a mouse strain expressing Cre under the control of a lineage-specific promoter (see Note 2). To precisely control Cre activity, use a CreERT2 or rtTA; tetO-Cre system which can be induced by administration of tamoxifen or doxycycline, respectively (for detailed methods on Cre-mediated recombination and genetic mouse reporter strains, see [8,9,10]). Use a ubiquitous promoter, such as the R26-CreERT2 mouse model [3, 12, 13], to stochastically label all cell lineages in the mammary gland in a more unbiased manner (see Note 3).
-
3.
To determine the contribution of a given cell from one lineage to the tissue, achieve clonal labeling density at the time of induction. Clonal labeling means that a sufficiently low number of cells is labeled to ensure that, at the time of analysis, all clones are truly clonal and represent the offspring of one labeled cell [9] (see Note 4).
-
4.
Determine the exact time points of initiation and termination of lineage tracing. Especially in the mammary gland, which undergoes several different phases of morphogenesis and remodeling, it is important to note that the clonal analysis will report the potency and dynamics of the labeled cells during the entire specified time frame of lineage tracing (see Note 5).
3.2 Mammary Gland Dissection
-
1.
Euthanize female mice at the desired time point in compliance with the institutional guidelines.
-
2.
Secure the mouse on a dissection pad with the ventral side upward and spray with 70% ethanol.
-
3.
Make a vertical incision through the skin along the midline using a pair of small scissors, extending from the lower abdomen to the upper chest area (Fig. 1, left panel).
-
4.
Separate the skin from the peritoneum using tweezers and secure the skin onto the dissection pad with pins.
-
5.
Locate the fat pads connected to the skin flaps on both sides, which contain the mammary epithelium (Fig. 1, right panel).
-
6.
To dissect the mammary glands, separate the complete fat pads from the skin.
-
7.
Gently grab the fat pad using blunt forceps and pull up (see Note 6). Remove the connective tissue and ligaments that attach the fat pad to the skin using a pair of fine dissection scissors.
-
8.
Collect the mammary glands (see Note 7) in a 50 mL falcon tube with 10 mL PBS on ice protected from light.
Schematic figure of the mammary gland dissection procedure. Left panel: A vertical midline incision through the skin, but leaving the peritoneum intact, will provide access to the subcutaneously located mammary glands. Right panel: Drawing of the location of the mammary glands after separating the skin from the peritoneum
3.3 Whole-Mount Preparation
-
1.
Dissolve collagenase A (2.5 mg/mL) and hyaluronidase (10 μg/mL) in PBS, and preheat this mild digestion solution at 37 °C.
-
2.
Transfer the mammary fat pads to a 12-well plate (see Note 8) and incubate in the collagenase A/hyaluronidase solution for 15–25 min (see Note 9) while shaking on an orbital shaker (100 rpm) at 37 °C.
-
3.
Wash the mammary glands 3 × 5 min in PBS while gently shaking.
-
4.
Prepare 10 mL of periodate-lysine-paraformaldehyde (PLP) buffer (see Note 10) in the meantime.
-
5.
Leave the PLP buffer at room temperature for 3–5 min and adjust the pH to 7.4 just before use (see Note 11).
-
6.
Incubate the mammary glands in PLP buffer in a 12- or 24-well plate and gently shake for 2 h at room temperature protected from light.
-
7.
Wash the mammary glands 3 × 5 min in PBS while gently shaking.
-
8.
Prepare blocking/permeabilization buffer. Mix all components well and incubate mammary glands for 3 h at room temperature while gently shaking on an orbital shaker.
-
9.
Replace the blocking/permeabilization buffer and add the desired primary antibody mix to the buffer (see Note 12). Incubate the mammary glands overnight in this mixture at room temperature while gently shaking.
-
10.
Wash the mammary glands 3 × 10 min in PBS with 0.2% Tween 20.
-
11.
Incubate the mammary glands with blocking/permeabilization buffer and add the desired secondary antibody mix to the buffer for at least 7 h at room temperature while gently shaking.
-
12.
Wash the mammary glands 3 × 10 min in PBS with 0.2% Tween 20. If desired, a nuclear stain such as DAPI or Hoechst could be added to the first washing step.
-
13.
Place each mammary gland on a coverslip (60 × 24 mm) and submerge in a few drops of anti-fade mounting medium (see Note 13). Spread the mammary gland onto the coverslip and avoid folds or twists of the tissue.
-
14.
Cover the mammary glands with another coverslip (60 × 24 mm) and gently push to flatten the tissue. Use sticky tape on both ends to keep the two coverslips together and seal the sides using nail polish (see Note 14).
-
15.
Keep the prepared whole-mount mammary glands at 4 °C and protected from light until further use.
3.4 Whole-Mount Confocal Imaging
-
1.
Place the whole mount in a sample holder for microscopy slides and fit into the automated stage of an inverted confocal microscope.
-
2.
The whole-mount preparation between two coverslips allows to image the specimen from both sides. Find the right focus using the oculars and determine the optimal side of the tissue for imaging (see Note 15). If the tissue is too thick or not translucent, it may be beneficial to image both sides of the whole mount, both images can be used later to reconstruct the ductal tree of the mammary gland.
-
3.
Determine the area of the whole-mount tissue and select a region of interest for a tile scan image (see Note 16). To perform quantitative analysis of the branched structure of the mammary gland, it is crucial to image the entire tissue and include all ducts and lobules.
-
4.
Determine the thickness of the tissue and set a Z-stack that includes all layers of the mammary epithelium. Depending on the amount of flattening of the tissue, a typical Z-stack covers 400 μm. A z-step size between 2 and 4 μm is recommended, with a minimum resolution of 512 × 512 pixels per tile (see Note 17).
-
5.
Use the desired laser lines to excite the endogenous and immunolabeled fluorophores. Depending on the amount and combination of fluorophores, a sequential imaging setting may be required to detect all fluorophores.
3.5 Image Processing
-
1.
Stitch the three-dimensional tile scan images together to generate a mosaic overview of the whole-mount mammary gland (Fig. 2a). Most imaging software packages have an automatic image stitching function built in.
-
2.
Perform qualitative analysis of the whole-gland images using volumetric rendering software (see Note 18), such as LASX 3D visualization (Leica Microsystems), Imaris (Bitplane AG), arivis (ZEISS), Amira (Visage Imaging), or other commercially available software packages.
-
3.
Export a 3D-rendered image or a 2D maximum intensity projection for all channels captured by confocal imaging (see Note 19). When using 3D-rendered images, make sure to export each channel in exactly the same orientation.
-
4.
Use the channel containing the signal of the immunolabeling with a ductal marker, for example, SMA staining to mark the basal cells or KRT8 staining for the luminal cells, to generate a clear image of the branched structure of the entire mammary gland. This can be achieved using the original image (if the quality is sufficient). Alternatively, thresholding can be used to reduce the background signal and generate a binary image of the ductal structure. Thresholding often leads to the loss of connections between ducts in the denser areas of the fat pad as a result of variations in immunolabeling intensity (see Note 20). When the ductal structure is dense or very complex, it is advised to generate a manual outline of the mammary ductal structure to prevent mistakes during later analysis steps (Fig. 3a, b).
-
5.
Map the number, location, and identity of the endogenously labeled fluorescent cells within the ductal tree as part of a (multicolor) lineage-tracing experiment (see Note 21). Several methods can be used depending on the quality of the acquired images. If the signal-to-noise ratio is good, a multicolor merged image can be used for quantification. However, often some regions of the mammary gland may contain more background signal and will benefit from signal correction or manual annotation. Using the cell-type-specific immunolabeling, cells of different subtypes can be identified.
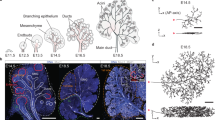
Whole-gland confocal imaging of the mammary gland. (a) Low-resolution overview tile scan of an entire fourth mammary gland derived from an R26-CreERT2het; R26-Confettihet female mouse. Clonal lineage tracing was initiated at the onset of puberty and analyzed at 8 weeks of age. The whole gland was labeled with an antibody against SMA. (b) Detailed confocal images of regions mapped back onto the low-resolution image in (a), indicated with colored boxes, showing clonal fragmentation (left panel), and cells of luminal and basal origin (right panels). Scale bars represent 5 mm (a), 100 μm (b, left panel), and 10 μm (b, right panels)
Whole-gland quantification pipeline. (a, b) Left: Confocal overview tile scan of a fourth mammary gland derived from an R26-CreERT2het; R26-Confettihet female mouse. Clonal lineage tracing was initiated at the onset of puberty and analyzed at 8 weeks of age. The whole gland was labeled with an antibody against KRT14. Right: To perform quantitative and clonal analysis, a schematic outline was generated based on the confocal images, and confetti cells were annotated within the ductal structure. (c) Screenshot of the branch analysis program before and after analysis. The approximate location of each branch point is indicated by a red dot
3.6 Branch Analysis and Clone Quantification
Several semiautomated branch analysis programs were developed to quantitatively describe the morphology of branched organs, such as the kidney [14, 15], the lung [16], or the mammary gland [17,18,19]. Most of these programs are based on skeletonization of the branched structure, which is subsequently used to derive features such as ductal area, perimeter, branch length distribution, and the number of branch points [17]. One commonly used program is tree surveyor, which is specifically built for optical projection tomography (OPT) datasets but can also be used to examine branched structures derived from other microscopy methods [7]. These programs are extremely useful to quantitatively assess many parameters of branched organs and can be used in combination with low-resolution whole-gland images derived from fluorescent samples imaged by OPT or light sheet microscopy or even images of H/E-stained samples.
To couple information on clone size, cell location, and cell identity to quantitative branch analysis data, aforementioned branch analysis programs are not suited. Therefore, we developed an intuitive branch analysis program which allows to quantitatively analyze the branched structure of multicolor .tif images combined with a function to map the location and number of luminal and basal cells within the branched structure of the mammary gland (Fig. 3c).
-
1.
Load the multicolor image of the whole-gland scan in the analysis program and indicate the origin of the tree (Fig. 3c, left panel).
-
2.
Activate the “Start Branch Point” button and draw a line to the next branch point by clicking on several points in the loaded image (Fig. 3c, middle panel). Click the “Start Branch Point” button again and a yellow dot will appear indicating the active branch point. Measure the width of the branch if desired using the “Width” button. Annotate all labeled cells within the annotated branch using the buttons on the right-hand side.
-
3.
Move to the next branch and repeat step 2 until the entire mammary gland is annotated (Fig. 3c, right panel). Active branch points are indicated with a yellow dot, and annotated branches will appear as red dots throughout the image.
-
4.
Once the branched structure is completely annotated, the output file (.txt) will contain all quantitative measurements to fully describe the branched structure of the annotated mammary gland, including branch length, branch width, number of branches, number of branch levels, and information on how branches are connected to each other (Fig. 4a). Moreover, for each branch, the location and identity of each annotated cell is stored as an integer along the length axis of the branch (Fig. 4a).
-
5.
The output data can be used to schematically rebuild the mammary gland (Fig. 4b, using the Ete2 Python toolkit for tree visualization) and perform quantitative analyses and modeling to understand how cellular dynamics drive branched tissue morphogenesis.
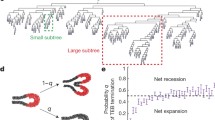
Quantitative analysis and reconstruction of the mammary gland (a) Screenshot of a typical branch analysis output file containing both information on the branching structure and the location, number, and identity of lineage-traced cells. (b) Whole-mammary gland plotted as a tree (top indicates the tree origin, with generation number increasing toward the bottom), showing large-scale branching heterogeneity with large and small subtrees at all generations. For each branch, the line indicates the color of the dominant clone (black indicates no Confetti+ cells present in the branch) and the pie chart indicates the ratio of each four colors labeled within the branch. Zooms on specific subtrees show the competition, separation, and loss of cell lineages over different branching levels in more detail. Regions 1 and 3 show evidence of clone fragmentation (the RFP clone in region 1 is fragmented in the left part of the tree. Similarly, the GFP clone in region 3 is fragmented throughout the entire subtree)
3.7 Modeling of Branching and Cellular Dynamics Based on Whole-Gland Image Analysis
From the datasets described above, a number of qualitative and quantitative features can be extracted and compared to different types of theoretical models. Below we provide a guide for the analysis of branching patterns (steps 1–5), followed by a description of the integration of clonal analysis data into these models (steps 6–7).
-
1.
Analyze the branch length distribution. Branch length is defined as the distance between two consecutive branching points. First verify that the branch length does not have specific dependencies with, for instance, branch generation number or distance from the branching origin (such dependency would result in nonmeaningful branch length distributions when grouping all branches together and would require binning the data in specific ways). Once this check is done, compare branch length distributions to different models for the stochasticity versus determinism of the branching process. For period branching models, such as Turing instabilities [20], one expects the branch length distribution to be highly peaked around a single well-defined value. For purely stochastic branching models [2], one expects a purely exponential distribution of branch length (i.e., long tails, and a wide variety of small and large branches).
-
2.
Analyze the subtree size distribution. Examine the distribution of subtree sizes—defined as the number of branches that emanate from a node at a given branch generation number—to distinguish stochastic versus deterministic models of branching (see Note 22). In contrast to the branch length distribution, the subtree size distribution can adopt a number of complex forms, which depend on the details of the model and the boundary conditions of the systems. Perform simple “branching and annihilating random walk” (BARW) simulations to build a theoretical expectation for this. The BARW model assumes that each tip can be described as a persistent random walk, which can (a) elongate with a certain speed v, leaving behind immobile ducts, (b) stochastically branch into two tips at any time with rate rb (so that the average branch length is proportional to v/rb), and (c) irreversibly terminate growth when reaching close proximity to another duct or the boundaries of the tissue (which can be arbitrarily defined based on the experimentally measured size of the organ and has the simplifying advantage of being nearly constant in time in mouse mammary gland). Run these simulations many times (typically 500–1000) to build precise computational subtree size distributions that can be compared to the experiments.
-
3.
Analyze the average spatial density of tips and ducts. Compute the number of duct and tip cells as a function of position along the axis of growth of the mammary gland, in both data and simulations. The BARW model predicts self-organized dynamical growth of the mammary tree at constant velocity, with a spatial “pulse” of growing tips at the invasion front, leaving behind a constant density of inactive ducts (see Note 23).
-
4.
Analysis of the spatial fluctuations in branching morphogenesis. Calculate the standard deviation in spatial density at different length scales (which provides theoretical insights into the branching process, see Note 24). This is done by “binning” the branching structure spatially at different length scales L and counting the average number N of cells (or alternatively branch length/area as a proxy) in a box of size L, as well as the associated standard deviation sN. Plotting the variance sN as a function of the average N (for many values of lengths L) can then serve as a metric for imperfection and nonuniformity of the tiling of space. Theoretically, one expects a power law sN ∝ N0.5 to correspond to minimal fluctuations and a power law sN ∝ N to correspond to maximal fluctuations (the entire mammary tree in one region of space while the rest of the fat pad is empty). Agreement between stochastic branching model and data, showing both intermediate scenarios of partially imperfect tiling, provides another qualitative and quantitative check for the model [2]. Alternatively, calculate the fractal dimension df of the branching structure [21], which increases with space-filling efficiency, and for branching in 2D must be between df = 1 (only a single, linear branch) and df = 2 (perfect space-filling from branching).
-
5.
Analysis of the branching angles and growth directionality. The angle of branches relative to the overall direction of growth of the organ is also a useful parameter to consider (see Note 25). Calculate the distribution of angles of each branch compared to the global direction of growth and compare it to different models of BARW with and without an external gradient locally guiding growing tips. Also calculate whether the angle of branching (between mother and daughter branches) changes as a function of generation number, local extracellular matrix fiber directionality, and/or repulsion between tips and ducts [21, 22], which can enhance overall growth directionality [23].
The advantage of developing a model for the branching morphogenesis of the entire organ is that this provides a backbone on which one can also simulate the dynamics and fate choices of the underlying stem cells/progenitors (see Note 26). At the cellular level, the mammary gland consists of basal and luminal cells populations, each divided into multiple sub-lineages [25]. Although at homeostasis quantitative lineage-tracing approaches can be used to understand the potency of each cell type [3, 26], the developmental situation is more complicated, as a given “clone” is not necessarily cohesive (or close to it as in homeostasis) but can span the entire tree (see Note 21). This is particularly problematic to assess multipotency, as, for instance, luminal and basal fragments of the same clone could be separated by large distances and thus be misinterpreted as two independent and unipotent clonal outcomes. Classical models of stem cell fate choices consider that a cell of type A has a given probability to self-renew (A → 2A) or differentiate into a different cell type (A → B) with associated stochastic rates and are simulated either without any reference to cell position (birth-death process) or on a fixed grid with division of a given cell influencing the fate of its nearest neighbors (voter-type model) [27]. These types of models can be generalized and coupled to the BARW models described above (see Note 27). Specific comparisons between model and data include (see steps 6 and 7):
-
6.
Analysis of cellular pluripotency during branching morphogenesis. Quantify the fraction of unipotent versus bipotent clones in the data. For very low clonal induction, the quantification is rather simple and does not per se require modeling if clones throughout the tree can be unambiguously reconstructed. However, for low to intermediate clonal induction, modeling of the simplest null hypothesis (pure unipotency of luminal and basal cell populations) is necessary to predict the fraction of “artificial” bipotency that emerges purely from a basal and luminal clone of the same color being independently labeled in a given tip. Compare the model to experimental fractions. Experimental bipotency significantly larger than the predicted “artificial” bipotency from the model demonstrates truly bipotent cells, which occurs, for instance, in early embryonic mammary development, but not in late embryonic and pubertal branching [3, 28, 29].
-
7.
Analysis of the number of functional stem cells per growing tip. Although the number of cells contained per growing tip can be counted by simple staining, this is not necessarily the number of functional stem cells responsible for the long-term growth of the tip (see Note 28). To estimate functional stem cell number, calculate the speed at which tips and ducts become monoclonal (i.e., consisting of a single-labeled color), which is inversely proportional to the number of functional stem cells at every tips (see Note 29). Via this type of quantitative comparisons between model and data, one can show, for instance, that pancreas branching morphogenesis relies on few functional stem cells [31], while nearly all cells in tips are functional stem cells for mammary gland [3].
4 Notes
-
1.
Most fluorescent reporter strains contain a stop codon flanked by LoxP sites in front of a sequence encoding one or more bright fluorophores, inserted behind a strong promoter sequence. Upon expression of Cre-recombinase, the stop-codon will be removed, resulting in irreversible expression of the fluorescent protein in the labeled cell and its progeny. A commonly used reporter strain is the multicolor R26-Confetti reporter [32, 33]. Upon Cre-mediated recombination, this reporter results in the stochastic expression of one of the four fluorophores it encodes (nuclear eGFP, YFP, RFP, or mCFP), with the advantage that more cells can be tracked at the same time at a clonal density within the same tissue structure.
-
2.
Some commonly used cell-type-specific promoters in the mammary gland are cytokeratin 14 (KRT14) [26, 34] or smooth muscle actin (SMA) [29, 35] for the basal lineage and cytokeratin 8 (KRT8) or Elf5 for the luminal lineage [34]. In addition to these more generic lineage promoters, a multitude of promoters has been used to trace cells of specific sub-lineages, including promoters of the estrogen receptor [36, 37], Lgr5 [38, 39], Lgr6 [40], Notch [41], or Axin2 [42].
-
3.
Ubiquitous induction is typically used to simultaneously determine the potency and contribution of any cell lineage during different developmental stages, such as pubertal development or pregnancy, in an unperturbed tissue setting, and often combined with a multicolor reporter strain.
-
4.
If the labeling density is too high, clones with the same fluorescent color may merge leading to erroneous quantitative analyses. Confetti reporters have the key advantage of providing a control to assess the clonality of the labeling: if all four colors have an equal 25% probability, then the number of non-clonal events can be estimated as long as “few” cells are labeled. Indeed, one can count the number of clones of different colors which are side by side (i.e., clones that would be grouped together if they were of the same color) and know probabilistically that the fraction of wrong groupings is to a third of this number. Taking YFP as an example, if two clones are labeled side by side, they can be YFP-YFP, YFP-RFP, YFP-GFP, or YFP-CFP with equal probability (again assuming all colors have 25% chance, although the argument can be adapted to more complicated cases [9]), so that the number of “undetectable” YFP-YFP events are a third of the number “detectable YFP-other color” events. For the mammary gland, this means that clonal labeling corresponds to a maximum of a few labeled cells per tip (i.e., approx. 1% labeling efficiency). Therefore, great care should be taken to titrate the amount of tamoxifen or doxycycline to achieve clonal labeling.
-
5.
For example, if lineage tracing is initiated at the onset of puberty, around 3 weeks of age, and mice are terminated after one round of pregnancy, the resulting clones report the contribution of these cells during puberty, some weeks of adulthood, as well as pregnancy, lactation, and involution. Importantly, during each of these stages, the labeled cell lineage may contribute differently, and as a result, it will be impossible to dissect the contribution of the labeled cell lineage during the different developmental stage, potentially leading to erroneous interpretation of the results. For instance, if the stage of interest is puberty, it will be important to initiate lineage tracing right before the onset of puberty at 3 weeks of age and perform clonal analysis at the end of puberty, but before the onset of the estrous cycle-driven remodeling of the mammary gland. Similarly, if the stage of interest is tissue turnover during adult homeostasis, the stage of the estrous cycle should be considered both during the initiation and termination of tracing. The formation of the alveolar buds during the estrous cycle can drastically affect the overall clone sizes observed in the mammary gland.
-
6.
It is important to not damage or cut through the fat pad, as this will result in an incomplete or damaged mammary epithelial tree.
-
7.
The fourth and fifth mammary glands are part of the same fat pad, and the second and third mammary glands are very close to each other. Therefore, the easiest is to dissect and process these two pairs of glands together.
-
8.
All steps should be performed while being protected from direct light. The best is to incubate the mammary glands in a 12- or 24-wells plate wrapped in aluminum foil.
-
9.
This incubation step results in mild digestion of the fat cells and collagen surrounding the mammary epithelium and is important for proper antibody penetration in the immune fluorescent staining step later on. The timing of this step depends on the size and thickness of the fat pad. It is crucial to check the glands every 5 min and stop the mild digestion when the first lipid droplets appear in the solution. Great care should be taken not to over-digest the tissue, as this will damage the mammary epithelium leading to potential disruption of the mammary ducts.
-
10.
PLP buffer is a mild fixative designed to retain endogenous fluorophores in thick tissues. Tissues fixated in PLP maintains tissue morphology and all (membrane) antigens for immunolabeling are well-preserved [43, 44].
-
11.
The pH after preparation is usually around 6.8, and 50–100 μl of NaOH is typically required to adjust the pH. Always prepare PLP buffer just before use, and do not use PLP buffer that is older than 1 day.
-
12.
To perform branching analysis, it is recommended to specifically immunolabel the mammary ductal epithelium, for example, by using antibodies against KRT14 or SMA labeling the basal cell layer or against E-cadherin or KRT8 labeling the luminal cell layer.
-
13.
If the tissue is not translucent after the staining procedure, an optional tissue clearing step could be used to improve visibility of the mammary epithelium during imaging. To prevent quenching of the (endogenous) fluorophores, aqueous clearing agents should be used [45, 46]. If the antigens for immunolabeling are preserved by the clearing method, it is advised to perform clearing prior to antibody staining.
-
14.
If the fat pad is very thick, a whole-gland mounting device could be used to further flatten the tissue (Fig. 5). This will improve visibility of the mammary epithelium within the fat pad.
-
15.
Depending on the desired resolution and time available, the best objective needs to be selected. To create a low-resolution overview of the entire mammary gland, a 10x dry objective can be used. To generate a more detailed map, in which individual cells and their identity (luminal or basal) can be readily recognized, a 20x magnification is minimally required. A higher magnification of 25x or 40x, combined with water or oil immersion, could be used to generate detailed imagines of regions of interest but is not recommended for large overview scans, because this will be time-consuming and generate large image files.
-
16.
Several microscope software programs have the option to generate a fast overview scan, which helps to determine the region of interest.
-
17.
A typical whole-mount scan (using a 20x dry objective) of an entire mammary gland with a cellular resolution typically takes 12–14 h using a regular confocal system, generating a file size of approximately 100 GB. To speed up the imaging process, faster microscopy technologies could be used, such as light sheet microscopy or OPT. These imaging technologies are more suited to capture thick tissue specimens and provide a good solution to solely look at the overall 3D morphology of intact tissues. However, cellular resolution is often not achieved if the tissue is thick and not fully transparent, and therefore these methods are not advised when one wants to combine both tissue morphology and cellular/clonal analyses.
-
18.
The 3D-rendered overview image of the mammary epithelial tree can serve as a road map to correlate high-resolution stacks, obtained with a 25x or 40x objective, to their coordinates within the entire ductal tree system (Fig. 2b).
-
19.
The mammary epithelium is a relatively flat tissue and the vast majority of the branches, with the exception of some side buds, extend into the xy-plane. Therefore, either 3D-rendered images or maximum intensity projections of the z-stacks can be used to perform branch analysis.
-
20.
Several methods have been described over the past years to correct for differences in intensity at different z-planes in branched structures [47, 48] which can improve the quality of a thresholded image.
-
21.
In most lineage-tracing experiments, cells are labeled at a clonal density leading to the outgrowth of local patches of cells with the same color after a defined period of lineage tracing. However, in a developmental setting such as the pubertal mammary gland, the tissue is expanding massively which can lead to clonal fragmentation. Using lineage tracing and whole-gland mapping, we have shown that stem/progenitor cells, located in the terminal end buds (TEBs) of the developing mammary gland, collectively proliferate to lay down the ductal tree [2, 3, 30]. If only one progenitor is labeled per TEB, only a small subset of cells will be labeled within the extending duct, intermingled with non-labeled offspring of the other progenitor cells. As a result, fragmented clones can be distributed over large parts of the ductal tree. Because of this phenomenon, it is important to generate whole-gland images to correctly assess the distribution and size for each clone during pubertal development.
-
22.
Generation number 1 is assigned to the branch at the origin of the mammary gland, with generation number increasing by 1 after each branching point. Such generation number can be defined unambiguously for simple trees without loops, which is the case for the mammary gland. In pubertal mammary branching, one can define this subtree origin to generation 5–6, as this represents the point at which branching starts on the onset of puberty [3]. For fractal branching patterns such as the lung, one expects well-defined subtree sizes, with distributions peaked again around a well-defined value, while for stochastically branching trees, one expects wide, long-tailed distributions.
-
23.
Predictions on the density and location of active tips can be verified experimentally by combining whole-tree reconstructions with short-term EdU incorporations assays (a 4 h pulse of EdU prior to dissection is typically enough to mark a large fraction of the proliferative cells in growing mammary tips) [2]. The location of the proliferative tips can be incorporated in the reconstruction procedure to give a proxy for the spatial density of active tips, and by averaging along the axis of mammary invasion, this can be directly compared to theory and simulations. The predictions from the BARW are not only computational as, unlike the one of subtree distribution, they can be derived by simple one-dimensional analytical theory [2].
-
24.
A key feature of the stochastic branching model described above is that although it can tile space in a robust and self-organized manner, it does so inefficiently, leaving behind large spatial fluctuations due to the stochastic nature of tip growth, which cannot be corrected later on due to the irreversible nature of tip termination in the model. This can be quantified, both in the model and in simulations, by several metrics such as variance of spatial fluctuations or fractal dimensions.
-
25.
Intuitively, if branches are strongly aligned toward the direction of growth, this could be a signature of external guidance in the system, for instance, if migrating cells in growing tips sense a pre-patterned external gradient guiding them (either chemical or mechanical). However, numerical simulations suggest that this is not necessarily the case. Firstly, it has been argued recently that migrating cells could also collectively generate their own directionality gradients, for instance, by consuming their own chemokine. As mentioned above, this can be modeled theoretically by adding tip-branch repulsion to the simple BARW model described above, although this has less impact than external guidance of the resulting branch directionality [21]. Secondly, it has been shown that even in the absence of repulsion, overall directionality could emerge in the BARW model. This can seem counterintuitive, as in this setting, tips perform a random walk in any direction with equal probability. However, because of tip termination, tips growing in the direction of overall invasion have more chance to “survive,” leading to an emergent, collective bias in angle directionality, which is relatively small compared to the ones generated by the gradients described above but still sufficient to explain the data.
-
26.
In principle, a comprehensive theory could encapsulate these multiple scales and explain from modeling individual cells and secreted morphogens how large-scale rules of branching emerge. However, in practice such multiscale theories are intractable, both mathematically, and because they contain too many adjustable parameters to make meaningful predictions. Thus, one should start by concentrating on each scale of description (molecular, cellular, organ) and adapt the modeling approach accordingly [24].
-
27.
These types of models can be generalized and coupled to BARW models, by considering that an active tip consists of N cells. Upon clonal induction, different clones divide and compete with each other inside of each tip, which can leave trails of ductal cells (proportionally to their prevalence inside a cell) and thus signatures of this competition. We can also model tip branching as a random partition of clonal cells from one to two tips, which will then each have independent growth and competition dynamics [3]. By assuming that proliferation in ducts is much slower than proliferation in tips (which is supported by data [3]), one can therefore see the clonal footprint in ducts as an “archaeological” record of the fate choices of tip cells, which can be compared between theory and data.
-
28.
For instance, if no intra-tip movements occur, one would expect the cells at the growing end of the tips to be in the most favored position and outcompete other cells of the tips via proliferation [30].
-
29.
For very few functional stem cells, one expects tips and ducts to rapidly become monoclonal (i.e., consisting of a single color), while for many functional stem cells, although the clone size is expected to increase linearly as a function of time/branch number because of clonal competition, it should do so very slowly.
Whole-gland mounting device. Photographs of the custom-made whole-gland mounting device in open and closed situations. The mounting device consists of a slot which fits a whole-mounted mammary gland bound by two coverslips. Place the lid on top of the mounted mammary gland and apply slight pressure by pushing down the levers. The whole-mount mammary gland can be kept in the mounting device until further use
References
Hogan BLM (1999) Morphogenesis. Cell 96:225–233
Hannezo E, Scheele CLGJ, Moad M et al (2017) A unifying theory of branching morphogenesis. Cell 171:242–255.e27. https://doi.org/10.1016/j.cell.2017.08.026
Scheele CLGJ, Hannezo E, Muraro MJ et al (2017) Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542:313–317. https://doi.org/10.1038/nature21046
Watson CJ, Khaled WT (2020) Mammary development in the embryo and adult: new insights into the journey of morphogenesis and commitment. Development 147. https://doi.org/10.1242/dev.169862
Maller O, Martinson H, Schedin P (2010) Extracellular matrix composition reveals complex and dynamic stromal-epithelial interactions in the mammary gland. J Mammary Gland Biol Neoplasia 15:301–318
Reed JR, Schwertfeger KL (2010) Immune cell location and function during post-Natal mammary gland development. J Mammary Gland Biol Neoplasia 15:329–339
Short K, Hodson M, Smyth I (2013) Spatial mapping and quantification of developmental branching morphogenesis. Development 140:471–478. https://doi.org/10.1242/DEV.088500
van de Moosdijk AAA, Fu NY, Rios AC et al (2017) Lineage tracing of mammary stem and progenitor cells. Methods Mol Biol 1501:291–308. https://doi.org/10.1007/978-1-4939-6475-8_15
Wuidart A, Ousset M, Rulands S et al (2016) Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev 30:1261–1277. https://doi.org/10.1101/gad.280057.116
Van Amerongen R (2015) Lineage tracing in the mammary gland using Cre/lox technology and fluorescent reporter alleles. Methods Mol Biol 1293:187–211. https://doi.org/10.1007/978-1-4939-2519-3_11
Roy E, Neufeld Z, Livet J, Khosrotehrani K (2014) Concise review: understanding clonal dynamics in homeostasis and injury through multicolor lineage tracing. Stem Cells 32:3046–3054. https://doi.org/10.1002/STEM.1804
Davis FM, Lloyd-Lewis B, Harris OB et al (2016) Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat Commun 7:13053. https://doi.org/10.1038/ncomms13053
Lloyd-Lewis B, Davis FM, Harris OB et al (2018) Neutral lineage tracing of proliferative embryonic and adult mammary stem/progenitor cells. Development 145. https://doi.org/10.1242/DEV.164079
Short KM, Combes AN, Lefevre J et al (2014) Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell 29:188–202. https://doi.org/10.1016/J.DEVCEL.2014.02.017
Combes AN, Short KM, Lefevre J et al (2014) An integrated pipeline for the multidimensional analysis of branching morphogenesis. Nat Protoc 912(9):2859–2879. https://doi.org/10.1038/nprot.2014.193
Metzger RJ, Klein OD, Martin GR, Krasnow MA (2008) The branching programme of mouse lung development. Nature 4537196(453):745–750. https://doi.org/10.1038/nature07005
Hadsell DL, Hadsell LA, Olea W et al (2015) In-silico QTL mapping of postpubertal mammary ductal development in the mouse uncovers potential human breast cancer risk loci. Mamm Genome 26:57–79. https://doi.org/10.1007/S00335-014-9551-X/TABLES/2
Blacher S, Gérard C, Gallez A et al (2016) Quantitative assessment of mouse mammary gland morphology using automated digital image processing and TEB detection. Endocrinology 157(4):1709–1716. https://doi.org/10.1210/en.2015-1601
Tolg C, Cowman M, Turley E (2018) Mouse mammary gland whole mount preparation and analysis. Bio-Protocol 8. https://doi.org/10.21769/BIOPROTOC.2915
Menshykau D, Blanc P, Unal E et al (2014) An interplay of geometry and signaling enables robust lung branching morphogenesis. Dev 141:4526–4536. https://doi.org/10.1242/dev.116202
Uçar MC, Kamenev D, Sunadome K et al (2021) Theory of branching morphogenesis by local interactions and global guidance. Nat Commun 12:1–10. https://doi.org/10.1038/s41467-021-27135-5
Prahl ALS, Viola JM, Liu J, Hughes AJ (2021) The developing kidney actively negotiates geometric packing conflicts to avoid defects. bioRxiv. https://doi.org/10.1101/2021.11.29.470441
Nerger BA, Jaslove JM, Elashal HE et al (2021) Local accumulation of extracellular matrix regulates global morphogenetic patterning in the developing mammary gland. Curr Biol 31:1903–1917.e6. https://doi.org/10.1016/j.cub.2021.02.015
Hannezo E, Simons BD (2019) Multiscale dynamics of branching morphogenesis. Curr Opin Cell Biol 60:99–105. https://doi.org/10.1016/j.ceb.2019.04.008
Twigger AJ, Khaled WT (2021) Mammary gland development from a single cell ‘omics view. Semin Cell Dev Biol 114:171–185. https://doi.org/10.1016/J.SEMCDB.2021.03.013
Van Keymeulen A, Rocha AS, Ousset M et al (2011) Distinct stem cells contribute to mammary gland development and maintenance. Nature 479:189. https://doi.org/10.1038/nature10573
Klein AM, Simons BD (2011) Universal patterns of stem cell fate in cycling adult tissues. Development 138:3103–3111. https://doi.org/10.1242/dev.060103
Wuidart A, Sifrim A, Fioramonti M et al (2018) Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol 20:666–676. https://doi.org/10.1038/s41556-018-0095-2
Lilja AM, Rodilla V, Huyghe M et al (2018) Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol 20:677. https://doi.org/10.1038/S41556-018-0108-1
Corominas-Murtra B, Scheele CLGJ, Kishi K et al (2020) Stem cell lineage survival as a noisy competition for niche access. Proc Natl Acad Sci 117:201921205. https://doi.org/10.1073/pnas.1921205117
Sznurkowska MK, Hannezo E, Azzarelli R et al (2018) Defining lineage potential and fate behavior of precursors during pancreas development. Dev Cell 46:360. https://doi.org/10.1016/j.devcel.2018.06.028
Snippert HJ, van der Flier LG, Sato T et al (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143:134–144. https://doi.org/10.1016/j.cell.2010.09.016
Livet J, Weissman TA, Kang H et al (2007) Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450:56–62. https://doi.org/10.1038/NATURE06293
Rios AC, Fu NY, Lindeman GJ, Visvader JE (2014) In situ identification of bipotent stem cells in the mammary gland. Nature 506:322–327. https://doi.org/10.1038/nature12948
Wendling O, Bornert JM, Chambon P, Metzger D (2009) Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47:14–18. https://doi.org/10.1002/DVG.20448
Van Keymeulen A, Fioramonti M, Centonze A et al (2017) Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep 20:1525–1532. https://doi.org/10.1016/j.celrep.2017.07.066
Wang C, Christin JR, Oktay MH, Guo W (2017) Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep 18:2825–2835. https://doi.org/10.1016/J.CELREP.2017.02.071
Plaks V, Brenot A, Lawson DA et al (2013) Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3:70–78. https://doi.org/10.1016/j.celrep.2012.12.017
Fu NY, Rios AC, Pal B et al (2017) Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat Cell Biol 19:164–176. https://doi.org/10.1038/ncb3471
Blaas L, Pucci F, Messal HA et al (2016) Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol 18:1346–1356. https://doi.org/10.1038/ncb3434
Rodilla V, Dasti A, Huyghe M et al (2015) Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol 13:e1002069. https://doi.org/10.1371/journal.pbio.1002069
Van Amerongen R, Bowman AN, Nusse R (2012) Developmental stage and time dictate the fate of Wnt/β-catenin- responsive stem cells in the mammary gland. Cell Stem Cell 11:387–400. https://doi.org/10.1016/j.stem.2012.05.023
Hall PA, Stearn PM, Butler MG, D’Ardenne AJ (1987) Acetone/periodate-lysine-paraformaldehyde (PLP) fixation and improved morphology of cryostat sections for immunohistochemistry. Histopathology 11(1):93–101
Pieri L, Sassoli C, Romagnoli P, Domenici L (2002) Use of periodate-lysine-paraformaldehyde for the fixation of multiple antigens in human skin biopsies. Eur J Histochem 46:365–375. https://doi.org/10.4081/1749
Lloyd-Lewis B, Davis FM, Harris OB et al (2016) Imaging the mammary gland and mammary tumours in 3D: optical tissue clearing and immunofluorescence methods. Breast Cancer Res 18:1–17. https://doi.org/10.1186/S13058-016-0754-9/TABLES/1
Rios AC, Capaldo BD, Vaillant F et al (2019) Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35:618–632.e6. https://doi.org/10.1016/J.CCELL.2019.02.010
Klingberg A, Hasenberg A, Ludwig-Portugall I et al (2017) Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J Am Soc Nephrol 28:452–459. https://doi.org/10.1681/ASN.2016020232/-/DCSUPPLEMENTAL
Østergaard MV, Sembach FE, Skytte JL et al (2020) Automated image analyses of glomerular hypertrophy in a mouse model of diabetic nephropathy. Kidney360 1(469–479):10.34067/KID.0001272019
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Authors
About this protocol
Cite this protocol
Hannezo, E., Scheele, C.L.G.J. (2023). A Guide Toward Multi-scale and Quantitative Branching Analysis in the Mammary Gland. In: Margadant, C. (eds) Cell Migration in Three Dimensions. Methods in Molecular Biology, vol 2608. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2887-4_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2887-4_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2886-7
Online ISBN: 978-1-0716-2887-4
eBook Packages: Springer Protocols