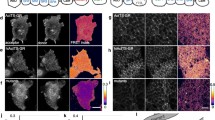
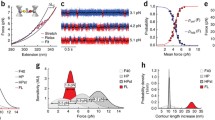
Abstract
The visualization of mechanical stress distribution in specific molecular networks within a living and physiologically active cell or animal remains a formidable challenge in mechanobiology. The advent of fluorescence-resonance energy transfer (FRET)-based molecular tension sensors overcame a significant hurdle that now enables us to address previously technically limited questions. Here, we describe a method that uses genetically encoded FRET tension sensors to visualize the mechanics of cytoskeletal networks in neurons of living animals with sensitized emission FRET and confocal scanning light microscopy. This method uses noninvasive immobilization of living animals to image neuronal β-spectrin cytoskeleton at the diffraction limit, and leverages multiple imaging controls to verify and underline the quality of the measurements. In combination with a semiautomated machine-vision algorithm to identify and trace individual neurites, our analysis performs simultaneous calculation of FRET efficiencies and visualizes statistical uncertainty on a pixel by pixel basis. Our approach is not limited to genetically encoded spectrin tension sensors, but can also be used for any kind of ratiometric imaging in neuronal cells both in vivo and in vitro.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Kung C (2005) A possible unifying principle for mechanosensation. Nature 436(7051):647–654
Katta S, Krieg M, Goodman MB (2015) Feeling force: physical and physiological principles enabling sensory mechanotransduction. Annu Rev Cell Dev Biol 31(1):347–371. https://doi.org/10.1146/annurev-cellbio-100913-013426
Venturini V, Pezzano F, Castro FC et al (2020) The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 370(6514). https://doi.org/10.1126/science.aba2644
Hoffman BD, Grashoff C, Schwartz MA (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475(7356):316–323
Krieg M, Dunn AR, Goodman MB (2015) Mechanical systems biology of C. elegans touch sensation. BioEssays 37(3):335–344. https://doi.org/10.1002/bies.201400154
Krieg M, Fläschner G, Alsteens D et al (2018) Atomic force microscopy-based mechanobiology. Nat Rev Phys 1:41–57. https://doi.org/10.1038/s42254-018-0001-7
Castro F, Venturini V, Ortiz-V’asquez S et al (2021) Direct force measurements of subcellular mechanics in confinement using optical tweezers. J Vis Exp 2021(174):1–35. https://doi.org/10.3791/62865
Gayrard C, Borghi N (2016) FRET-based molecular tension microscopy. Methods 94(2016):33–42. https://doi.org/10.1016/j.ymeth.2015.07.010
Hurst S, Vos BE, Brandt M et al (2021) Intracellular softening and increased viscoelastic fluidity during division. Nat Phys 17(11):1270–1276. https://doi.org/10.1038/s41567-021-01368-z
Grashoff C, Hoffman BD, Brenner MD et al (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466(7303):263–266
Canever H, Carollo PS, Fleurisson R et al (2021) Molecular tension microscopy of E-Cadherin during epithelial- mesenchymal transition. Methods Mol Biol 2179:289–299
Krieg M, Dunn AR, Goodman MB (2014) Mechanical control of the sense of touch by β-spectrin. Nat Cell Biol 16(3):224–233. https://doi.org/10.1038/ncb2915
Cai D, Chen SC, Prasad M et al (2014) Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157(5):1146–1159. https://doi.org/10.1016/j.cell.2014.03.045
Vuong-Brender TTK, Boutillon A, Rodriguez D et al (2018) HMP-1/ α-catenin pro- motes junctional mechanical integrity during morphogenesis. PLoS ONE 13(2):1–21. https://doi.org/10.1371/journal.pone.0193279
Ye AA, Cane S, Maresca TJ (2016) Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat Commun 7:1–9. https://doi.org/10.1038/ncomms13221
Lemke SB, Weidemann T, Cost AL et al (2018) A small proportion of Talin molecules transmit forces to achieve muscle attachment in vivo. PLoS Biol 310(939):446336. https://doi.org/10.1101/446336
Conway DDE, Breckenridge MMT, Hinde E et al (2013) Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol 23(11):1024–1030. https://doi.org/10.1016/j.cub.2013.04.049.Fluid
Aird EJ, Tompkins KJ, Ramirez MP et al (2020) Enhanced molecular tension sensor based on Bioluminescence Resonance Energy Transfer (BRET). ACS Sensors 5(1):34–39. https://doi.org/10.1021/acssensors.9b00796
Ringer P, Weißl A, Cost AL et al (2017) Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1. Nat Methods 14(11):1090–1096. https://doi.org/10.1038/nmeth.4431
Paszek MJ, DuFort CC, Rossier O et al (2014) The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511(7509):319–325
Guo J, Wang Y, Sachs F et al (2014) Actin stress in cell reprogramming. Proc Natl Acad Sci 111(49):E5252–E5261. https://doi.org/10.1073/pnas.1411683111
Meng F, Suchyna TM, Sachs F (2008) A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J 275(12):3072–3087
Ye N, Verma D, Meng F et al (2014) Direct observation of α-actinin tension and recruitment at focal adhesions during contact growth. Exp Cell Res 327(1):57–67. https://doi.org/10.1016/j.yexcr.2014.07.026
LaCroix AS, Lynch AD, Berginski ME et al (2018) Tunable molecular tension sensors reveal extension-based control of vinculin loading. elife 7:1–36. https://doi.org/10.7554/elife.33927
Arsenovic PT, Ramachandran I, Bathula K et al (2016) Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J 110(1):34–43. https://doi.org/10.1016/j.bpj.2015.11.014
D́ejardin T, Carollo PS, Sipieter F et al (2020) Nesprins are mechanotransducers that discriminate epithelial-mesenchymal transition programs. J Cell Biol 219(10). https://doi.org/10.1083/JCB.201908036
Borghi N, Sorokina M, Shcherbakova OG et al (2012) E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A 109(31):12568–12,573
Price AJ, Cost AL, Ungewiß H et al (2018) Mechanical loading of desmosomes depends on the magnitude and orientation of external stress. Nat Commun 9(1). https://doi.org/10.1038/s41467-018-07523-0
Sanfeliu-Cerdán N, Mateos B, Garcia-Cabau C et al (2022) A rigidity phase transition of Stomatin condensates governs a switch from transport to mechanotransduction. BioRxiv. https://doi.org/10.1101/2022.07.08.499356
Brenner MD, Zhou R, Conway DE et al (2016) Spider silk peptide is a compact, linear nano-spring ideal for intracellular tension sensing. Nano Lett 16(3):2096–2102. https://doi.org/10.1021/acs.nanolett.6b00305
Meng F, Sachs F (2012) Orientation-based FRET sensor for real-time imaging of cellular forces. J Cell Sci 125(3):743–750
Elangovan M, Wallrabe H, Chen Y et al (2003) Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods 29(1):58–73
Day RN, Booker CF, Periasamy A (2008) Characterization of an improved donor fluorescent protein for Forster resonance energy transfer microscopy. J Biomed Opt 13(3):31203
LaCroix AS, Rothenberg KE, Berginski ME et al (2015) Construction, imaging, and analysis of FRET-based tension sensors in living cells, vol 125. Elsevier Ltd. https://doi.org/10.1016/bs.mcb.2014.10.033
Cost AL, Ringer P, Chrostek-Grashoff A et al (2015) How to measure molecular forces in cells: a guide to evaluating genetically-encoded FRET-based tension sensors. Cell Mol Bioeng 8(1):96–105. https://doi.org/10.1007/s12195-014-0368-1
Das R, Lin LC, Catala-Castro F et al (2021) An asymmetric mechanical code ciphers curvature-dependent proprioceptor activity. Sci Adv 7:eabg4617. https://doi.org/10.1126/sciadv.abg4617
Kelley M, Yochem J, Krieg M et al (2015) FBN-1, a fibrillin-related protein, is required for resistance of the epidermis to mechanical deformation during c. elegans embryogenesis. elife 2015(4):1–71. https://doi.org/10.7554/eLife.06565
Johnson CP, Tang HY, Carag C et al (2007) Forced unfolding of proteins within cells. Science (New York, NY) 317(5838):663–666
Gates EM, LaCroix AS, Rothenberg KE et al (2019) Improving quality, reproducibility, and usability of FRET-based tension sensors. Cytometry Part A 95(2):201–213. https://doi.org/10.1002/cyto.a.23688
Fischer LS, Rangarajan S, Sadhanasatish T et al (2021) Molecular force measurement with tension sensors. Annu Rev Biophys 50:595–616. https://doi.org/10.1146/annurev-biophys-101920-064756
Sands B, Burnaevskiy N, Yun SR et al (2018) A toolkit for DNA assembly, genome engineering and multicolor imaging for C. elegans. Transl Med Aging 2(2018):1–10. https://doi.org/10.1016/j.tma.2018.01.001
Porta-de-la Riva M, Fontrodona L, Villanueva A et al (2012) Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp 64:e4019. https://doi.org/10.3791/4019
Kim E, Sun L, Gabel CV et al (2013) Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS ONE 8(1):e53419
Bao Z, Murray JI (2011) Mounting Caenorhabditis elegans embryos for live imaging of embryogenesis. Cold Spring Harb Protoc 6(9):1089–1094. https://doi.org/10.1101/pdb.prot065599
Rivera Gomez KA, Schvarzstein M (2018) Immobilization nematodes for live imaging using an agarose pad produced with a Vinyl Record. microPublication Biology. https://doi.org/10.17912/QG0J-VT85
Jeong B, Kim SW, Bae YH (2012) Thermosensitive sol-gel reversible hydrogels. Adv Drug Deliv Rev 64(SUPPL):154–162. https://doi.org/10.1016/j.addr.2012.09.012
Gilleland CL, Rohde CB, Zeng F et al (2010) Microfluidic immobilization of physiologically active Caenorhabditis elegans. Nat Protoc 5(12):1888–1902
Burnett K, Edsinger E, Albrecht DR (2018) Rapid and gentle hydrogel encapsulation of living organisms enables long-term microscopy over multiple hours. Commun Biol 1(1). https://doi.org/10.1038/s42003-018-0079-6
Kopito RB, Levine E (2014) Durable spatiotemporal surveillance of Caenorhabditis elegans response to environmental cues. Lab Chip 14(4):764–770. https://doi.org/10.1039/c3lc51061a
Wen Q, Po MD, Hulme E et al (2012) Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron 76(4):750–761
Lockery SR, Lawton KJ, Doll JC et al (2008) Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J Neurophysiol 99(6):3136–3143. https://doi.org/10.1152/jn.91327.2007
Lambert TJ (2019) FPbase: a community-editable fluorescent protein database. Nat Methods 16(4):277–278. https://doi.org/10.1038/s41592-019-0352-8
Chen H 3rd, Koushik SV et al (2006) Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys J 91(5):L39–L41
Sun Y, Periasamy A (2010) Additional correction for energy transfer efficiency calculation in filter-based Förster resonance energy transfer microscopy for more accurate results. J Biomed Opt 15(2):020513. https://doi.org/10.1117/1.3407655
Feige JN, Sage D, Wahli W et al (2005) PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc Res Tech 68(1):51–58
Koushik SV, Chen H, Thaler C et al (2006) Cerulean, Venus, and VenusY67C FRET reference standards. Biophys J 91(12):L99–L101
Esposito A (2020) How many photons are needed for FRET imaging? Biomed Opt Express 11(2):1186. https://doi.org/10.1364/boe.379305
Hulme SE, Shevkoplyas SS, Apfeld J et al (2007) A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab Chip 7(11):1515
Berger S, Spiri S, DeMello A et al (2021) Microfluidic-based imaging of complete Caenorhabditis elegans larval development. Development (Cambridge) 148(18). https://doi.org/10.1242/DEV.199674
Heintzmann R (2006) Band limit and appropriate sampling in microscopy. Cell Biol 3:29–36. https://doi.org/10.1016/B978-012164730-8/50131-3
Kenworthy AK, Edidin M (1998) Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J Cell Biol 142(1):69–84
Morris C, Foster OK, Handa S et al (2018) Function and regulation of the Caenorhabditis elegans Rab32 family member GLO-1 in lysosome-related organelle biogenesis. PLoS Genet 14(11):1–36. https://doi.org/10.1371/journal.pgen.1007772
Datta R, Heaster TM, Sharick JT et al (2020) Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J Biomed Opt 25(07):1. https://doi.org/10.1117/1.jbo.25.7.071203
Meiresonne NY, Consoli E, Mertens LM et al (2019) Superfolder mTurquoise2 ox optimized for the bacterial periplasm allows high efficiency in vivo FRET of cell division antibiotic targets. Mol Microbiol 111(4):1025–1038. https://doi.org/10.1111/mmi.14206
Costantini LM, Baloban M, Markwardt ML et al (2015) A palette of fluorescent proteins optimized for diverse cellular environments. Nat Commun 6(May). https://doi.org/10.1038/ncomms8670
Arsenovic PT, Mayer CR, Conway DE (2017) SensorFRET: a standardless approach to measuring pixel-based spectral bleed-through and FRET efficiency using spectral imaging. Sci Rep 7(1):1–15. https://doi.org/10.1038/s41598-017-15,411-8
Acknowledgments
We thank the ICFO SLN facility for the generous use of their microscopes. M.K. acknowledges financial support from the Spanish Ministry of Economy and Competitiveness through the Plan Nacional (PGC2018-097882-A-I00), “Severo Ochoa” program for Centres of Excellence in R&D (CEX2019-000910-S; RYC-2016-21062), from Fundació Privada Cellex, Fundació Mir-Puig, and from Generalitat de Catalunya through the CERCA and Research program (2017 SGR 1012), in addition to funding through ERC (MechanoSystems) and HFSP (CDA00023/2018), la Caixa Foundation (ID 100010434, LCF/BQ/DI18/11660035), and MINECO (FPIPRE2019-088840 funded by MCIN/AEI/10.13039/501100011033 and ESF “Investing in your future” to N.S.). M.B.G. is supported by NIH Grant R35105092 and A.R.D. by NIH grant 1R35GM130332-01.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Sanfeliu-Cerdán, N., Lin, LC., Dunn, A.R., Goodman, M.B., Krieg, M. (2023). Visualizing Neurons Under Tension In Vivo with Optogenetic Molecular Force Sensors. In: Zaidel-Bar, R. (eds) Mechanobiology. Methods in Molecular Biology, vol 2600. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2851-5_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2851-5_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2850-8
Online ISBN: 978-1-0716-2851-5
eBook Packages: Springer Protocols