Abstract
Understanding brain function requires technologies that can monitor and manipulate neural activity with cellular resolution and millisecond precision in three dimensions across large volumes. These technologies are best designed using interdisciplinary approaches combining optical techniques with reporters and modulators of neural activity. While advances can be made by separately improving optical resolution or opsin effectiveness, optimizing both systems together matches the strengths and constraints of different approaches to create a solution optimized for the needs of neuroscientists. To achieve this goal, we first developed a new multiphoton photoexcitation method, termed 3D-Scanless Holographic Optogenetics with Temporal focusing (3D-SHOT), that enables simultaneous photoactivation of arbitrary sets of neurons in 3D. Our technique uses point-cloud holography to place multiple copies of a temporally focused disc, matched to the dimensions of a neuron’s cell body, anywhere within the operating volume of the microscope. However, since improved placement of light, on its own, is not sufficient to allow precise control of neural firing patterns, we also developed and tested optogenetic actuators ST-ChroME and ST-eGtACR1 that fully leverage the new experimental capabilities of 3D-SHOT. The synergy of fast opsins matched with our technology allows reliable, precisely timed control of evoked action potentials and enables on-demand read-write operations with unprecedented precision. In this chapter, we review the steps necessary to implement 3D-SHOT and provide a guide to selecting ideal opsins that will work with it. Such collaborative, interdisciplinary approaches will be essential to develop the experimental capabilities needed to gain causal insight into the fundamental principles of the neural code underlying perception and behavior.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Optogenetics
- Temporal focusing
- 3D holography
- ChroME
- Brain-machine interfacing
- Multiphoton
- 3D-SHOT
- Soma-targeted
- Opsin
1 Introduction
Although optogenetics have become a mainstay of neuroscience research, used to probe causal relationships between circuit activity and behavior [1,2,3,4,5,6], it is only recently that multiphoton optogenetic techniques have been used to modulate neural activity. Numerous technical advances in optics [7,8,9,10] and opsins [11,12,13,14,15] over the last decade have led to an increase in usage and adoption of multiphoton optogenetic strategies. Multiphoton optogenetics have been used to examine visual discrimination [3, 14], coding features of detection [16, 17], ensemble activity [18], cortical circuitry [19], and more [20, 21].
In this chapter, we will briefly review the state of the field and introduce 3D-SHOT [7]. We will detail the steps required to build, align, calibrate, and validate 3D-SHOT as an add-on on the light path of a 2-photon microscope. We will then discuss how to assess and select ideal opsin and reporter combinations for use with a 3D-SHOT system.
Optogenetic techniques have been rapidly and widely adopted in neuroscience research because they enable precise and reversible external control of neural activity with high temporal precision by means of minimally invasive optical signals. However, the spatial precision is generally too poor to manipulate individual neurons because light does not propagate well through dense brain tissue. The vast majority of optogenetics studies primarily leverage genetic specificity rather than spatial control. However, since many neural computations and behaviors rely on populations of neurons that are genetically similar but spatially intermixed [18, 22,23,24], precise targeting of individual neurons with optical methods is necessary. Two-photon activation of opsins is an attractive approach for improving spatial resolution. The longer wavelengths used in two-photon excitation are less affected by optical scattering [25], which dramatically improves the axial resolution and the accessible depth of sculpted illumination patterns [26, 27]. Further, two-photon absorption is a nonlinear effect which further restricts opsin excitation to a narrow axial plane [8, 28].
Most biological studies using 2-photon optogenetics have used scanning-based approaches [3, 14, 18,19,20,21]. Similar in principle to two-photon imaging, a femtosecond-pulsed infrared laser beam is focused into a single diffraction-limited spot which is scanned in 2D or 3D, with galvo-mirrors [27], acousto-optic modulators [29, 30], spatial light modulators (SLM) [31], or micro-electromechanical systems (MEMS) [32]. A raster [8, 13] or spiral [18, 28, 33] pattern is scanned across the soma to target neurons in 3D. These techniques are power efficient [34] but require opsins that are either extremely strong or slow to deactivate (ideally both), so that the photocurrent can accumulate as the spot is scanned across the neuron soma, usually at the expense of temporal resolution [11].
To improve temporal precision, whole-cell illumination techniques that forgo scanning have been developed to simultaneously illuminate the entire cell soma with a larger spot and activate all the opsin at once. Some approaches achieve whole-cell activation with low NA objective [13, 20] at the expense of spatial resolution, but the preferred method relies on computer-generated holography (CGH) [35,36,37] with a spatial light modulator (SLM) to synthesize custom illumination patterns that are matched to the shape of individual neurons (see also Chaps. 3 and 11). Compared to scanning approaches, whole-cell activation with CGH enables faster responses to optogenetic stimulation, but requires higher peak powers. With traditional multiphoton CGH [38], and even point scanning methods [33], spatial resolution along the optical axis is determined by how rapidly the power density is attenuated as light propagates into and out of the targeted area. These often result in significant but undesired photoexcitation above and below the target. In practice, physiological spatial resolution is highly power-dependent, and single neuron spatial resolution (e.g., axial FWHM ~30 μm) [9, 39] is generally impossible across large volumes, even with high numerical aperture (NA) objectives.
To eliminate the tradeoff between target dimensions in the (x,y) plane and decreased axial (z) resolution [40, 41], 3D-SHOT relies on temporal focusing (TF) [40, 41] where a diffraction grating decomposes femtosecond pulses into separate colors, such that the different wavelengths components within the original pulse propagate along separate light paths. Each component of the decomposed pulse has a narrow spectral bandwidth and is therefore broadened in time, which dramatically reduces the peak intensity and prevents two-photon absorption. However, two-photon absorption can be enabled again when the original pulse is retrieved by constructive interference of all the chromatically separated components at conjugate images of the diffraction grating [40, 41]. TF restricts multiphoton absorption to a narrow (z) depth that depends on the grating’s spatial frequency, not on the dimensions of the targeted area. TF has been successfully applied for selective two-photon tomographic fluorescence imaging [42, 43], has been implemented with mechanical scanning [44], and with random-access volume sampling of functional fluorescence [45]. A detailed presentation of TF is available in Chap. 9.
For two-photon photostimulation applications, TF can activate opsins over a wide area matching the neuron’s shape in the focal plane, without compromising depth specificity [39, 46]. TF also mitigates scattering [47, 48] even through thick layers of brain tissue [49, 50] (see also Chap. 9). Although multiphoton CGH with TF can achieve wide-field photostimulation with cellular resolution and high temporal precision, most implementations only enable excitation within a single 2D plane [20, 39, 46]. Thus, neurons located above or below the focal plane are not addressable, a necessary condition for many experiments designed to interface with neural circuits in vivo, where relevant neurons may be localized anywhere in the 3D volume of interest. Multi-level temporal focusing has been implemented with holograms tiled into clusters on the SLM surface which can be individually defocused in space by applying digital lens patterns on a second SLM [51]. This strategy is limited in the number of distinct depth levels used before in-plane resolution is degraded, constraining the neuronal population that is simultaneously addressable with optical stimuli.
To overcome this outstanding challenge 3D-SHOT leverages the advantages of CGH to simultaneously address custom 3D locations on demand, and TF for enhanced spatial resolution at the scale of individual neuron soma. 3D-SHOT forgoes the ability to create custom patterns to make TF and 3D CGH compatible, instead it replicates multiple identical copies of a temporally focused excitation pattern matched to the dimensions of a neuron soma at each target location. The result is a technology that is specifically tailored to optogenetic photostimulation applications and enables single-shot in vivo photoactivation of custom neuron ensembles distributed anywhere in the accessible volume, with single-neuron spatial resolution.
Understanding the causal relationship between neural activity and behavior is a fundamental goal of neuroscience. However, this causal inference requires manipulations that act at the scale of natural activity, i.e., writing temporally precise patterns of activity in many cells with single-cell specificity. While optical approaches such as spiral scanning or 3D-SHOT can confine light to the dimensions of a single cell, molecular actuators, also known as opsins, are required to convert light into neural activity. The properties of these opsins and how they interact with the stimulation system will determine how well one can drive precise trains of activity. The fast and potent opsin ChroME [11] was engineered alongside the development of 3D-SHOT to achieve this goal, but is only a single example of a class of opsins with appropriate speed and sensitivity properties that have yet to be discovered. As such, the fourth section of this chapter outlines the criteria necessary to select an opsin optimally paired to 3D-SHOT for the purpose of writing precise trains of activity into groups of neurons.
Fundamental to the goal of writing temporally specific patterns is to pair extremely fast opsins with “flash”-based optical approaches, i.e., those that simultaneously illuminate an entire cell such as 3D-SHOT. In order to drive precisely timed action potentials faithfully and at high rates, the underlying evoked photocurrents must be very strong and very fast – both rising and falling very rapidly. Simultaneous illumination systems ensure that all possible opsin molecules are activated at the same time, leading to the shortest response time, while opsins with both fast kinetics and high conductance ensure that action potentials are faithfully driven, without the risk of creating doublets. Indeed, opsins with slower decay kinetics tend to have higher overall conductance but are not capable of driving pyramidal cells at high rates and often have large jitter in action potential timing [34, 52, 53].
Finally, multiphoton optogenetic stimulation is rarely performed as a standalone technique, and is almost always paired with calcium imaging. Special considerations are needed to match the properties of opsins and reporters to ensure that both multiphoton optogenetics and imaging are compatible for simultaneous use. At present, the most successful imaging approaches rely on “green” calcium indicators (i.e., the GCaMP series) that absorb ~920 nm matched to “red” opsins with longer excitation wavelengths. However, even opsins with peak absorption >1000 nm are sensitive to these imaging wavelengths. We discuss these constraints and the mitigation approaches that are available to combine multiphoton optogenetics with imaging under those circumstances.
Precise control of neural activity cannot be achieved by any one technique alone. New optical developments, new molecular tools, and new approaches to unite these devices are needed to reach the next step of precise causal manipulation of neural systems. As technologies are constantly being developed and improved; new experimental capabilities provide a path to answer previously intractable neuroscience questions. By combining different skillsets and expertise, the technological solutions that emerge are better than what would have been created from the perspective of a single discipline. Our intent is for this chapter to serve as a guide and resource for future users who will implement and improve upon multiphoton optogenetics and usher in a new epoch of neuroscience discovery.
2 Methods
2.1 3D-SHOT Optical System Design
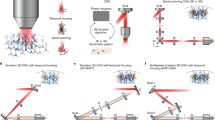
Our experimental setup, shown in Fig. 1, is based on the standard design of a holographic microscope with a spatial light modulator (SLM) in the Fourier domain (pupil plane). The SLM shapes the phase of a coherent femtosecond laser light source to synthesize custom 3D shapes [54, 55] digitally synthesized with Computer Generated Holography [35,36,37] (CGH). Unlike scanning approaches, CGH wide-area holograms matched to the dimensions of each neuron’s soma enable simultaneous, flash-based, activation of a large number of opsin molecules, yielding photocurrents with fast kinetics [39].
Experimental setup for 3D-SHOT. This is made of two consecutive optical systems. First, a diffraction grating and a rotating diffuser are imaged onto each other by a f-f optical relay. This assembly shapes femtosecond laser pulses both spatially and spectrally to create a custom temporally focused pattern (CTFP) matched to the dimensions of a neuron soma. The resulting engineered point spread function is then spatially modulated by a second system that enables 3D computer-generated holography (CGH). A spatial light modulator (SLM) placed in the Fourier domain modulates the phase of the CTFP to target custom 3D locations with a point-cloud hologram. The resulting sculpted illumination pattern replicates identical copies of the CTFP at each targeted location. The 3D hologram is further demagnified by a tube lens and a microscope objective. A zero-order block eliminates any remaining undiffracted light from the hologram. The grating frequency determines the spectral dispersion, “a”, and the diffuser determines the beam dimension “b” at the surface of the SLM. Those parameters, along with the focal lengths of lenses L1–L4, are adjusted to match the desired addressable volume and CTFP dimensions within constraints imposed by the SLM size, the laser source, and the numerical aperture of the microscope
In most brain structures, neurons are distributed continuously in 3D, not in discrete layers. Therefore, the inability of 2D optogenetics approaches (i.e., 2D CGH with temporal focusing) to target neurons at any arbitrary number of axial planes simultaneously is a major obstacle for large-scale optogenetic interrogation of neural circuits. To overcome this outstanding challenge 3D-SHOT leverages the advantages of 3D-CGH, to simultaneously address neurons in custom locations. To make 3D CGH and temporal focusing mutually compatible, 3D-SHOT forgoes the ability to create custom patterns. Instead, the optical path is optimized to holographically replicate multiple identical copies of a temporally focused excitation pattern, termed “custom temporally focused pattern” (CTFP). The CTFP is specifically engineered to match the dimensions of a neuron’s soma, and to be compatible with 3D CGH so that identical copies of the CTFP, with individually specified brightness can be placed at each target neuron anywhere in the accessible 3D volume. The result is a technology that is tailored for optogenetics applications and enables single shot in vivo photoactivation of custom neuron ensembles distributed anywhere in the accessible volume, with single-neuron spatial resolution.
To implement 3D SHOT in a multiphoton microscope, the first step is to create a static, temporally focused object matched to the dimensions of the desired target. For this, we illuminate a reflective blazed diffraction grating with a femtosecond laser light source. The incidence angle is adjusted so that the first diffracted order reflects orthogonally to the surface of the grating. The grove depth, material, coating, and incoming wave polarization are adjusted to best match the desired LASER wavelength and optical power density and to maximize the amount of light in the first diffracted order. Lenses L1 and L2 are in a 4-f configuration and create an exact optical relay that place a virtual copy of the diffraction grating at the surface of the transparent, rotating diffuser. The rotating diffuser applies an engineered (gaussian) phase pattern to the temporally focused image that is continuously randomized by the mechanical rotation. This phase perturbation is necessary to distribute the energy in the Fourier domain (i.e., to uniformly illuminate the SLM), a critical step that enables the compatibility between 3D CGH and temporal focusing and maximizes the diffraction efficiency.
The CTFP is matched to the characteristic dimensions of a neuron soma by selecting the magnification M = f2/f1, where f2 and f1 designate the focal lengths of lenses L2 and L1 respectively. The axial confinement (temporal focusing) can be adjusted by selecting diffraction gratings with a higher (or lower) grating spatial frequency (in lines per mm). Both properties can be adjusted independently of the additional phase perturbation induced by the rotating diffuser.
A second 4-f system made with lens L3 and L4 with focal length f3, resp. f4 relays the CTFP, first to the SLM in the pupil plane, then to the volume where the 3D hologram is first synthesized. In this configuration, the SLM applies a multiplicative phase pattern in the Fourier domain, which corresponds to a convolution operation in the real domain. To utilize 3D SHOT for neural stimulation, a CGH algorithm only needs to compute a hologram made of a 3D cloud of diffraction-limited points centered on each target neuron, and the optical system will yield one copy of the CTFP at each target point. To compensate for spatially dependent diffraction efficiency, and non-uniform losses through the optical system, the respective brightness of each target can be adjusted by computing digitally compensated holograms that redistribute the available laser intensity across each point of the 3D cloud based on expected losses, and the total energy can be adjusted globally across all targets by modulating the power of the laser beam.
2.1.1 3D-SHOT Design Parameters
-
1.
The size of the CTFP must be adjusted to match the dimensions “d” of a neuron. In a typical implementation of 3D SHOT, the 3D hologram (see Fig. 1) is relayed into a microscope with an additional tube lens (L5, f = f5) and microscope objective (L6, f = f6) with magnification M = f5/f6. The dimensions of the CTFP is M·d in the 3D hologram, Mdf3/f4 at the rotating diffuser, and correspond to an incoming beam of width M·d·f3 f1/(f4f2) at the blazed grating.
-
2.
The rotating holographic diffuser is a transparent material with an engineered surface that deflects incoming light in a Gaussian pattern angular distribution with a characteristic diffraction angle, αd. The diffuser spatially stretches the wave in the Fourier domain by an amount b (see Fig. 1), optimized to ensure an even illumination of the SLM active area, and given by.
-
3.
The line spacing of the blazed diffraction grating, l, or spatial frequency, fg, (l = 1/fg) must be adjusted to match the spectral bandwidth δλ of the femtosecond laser, and the desired dimensions of the CTFP along the (z) axis. The angular dispersion of the diffraction grating is given by α = δλ/l, and stretches the pulse in the Fourier domain. At a distance f1 from lens L1, the dimension of the stretched pulse satisfies af2/f3 = α f1. Hence, the spectral dispersion, a, at the SLM, is given by:
The numerical aperture NA of the microscope objective is generally the limiting factor that defines the accessible volume and CTFP minimal dimensions for 3D-SHOT. The SLM pattern (see Fig. 1) of width a + 2b is imaged onto the back aperture of the microscope objective. Hence, to fully capture the light modulated by the SLM, a suitable design constraint is to ensure (a + 2b)f5/f4 < NA f6. In this configuration, the characteristic size, δz, of the CTFP along the (z) axis in the demagnified hologram under the microscope objective is given by
2.1.2 Implementation Guidelines for 3D-SHOT
3D-SHOT is implemented as a secondary system on the path of a multiphoton microscope that is also designed for two-photon imaging, typically with a secondary laser. Imaging and photostimulation are most efficiently merged with a dichroic mirror or a polarizing beam splitter cube. When using a polarizing cube, the orientation of the SLM, grating, and path-merging cube must be adjusted to match polarization constraints, with one path (e.g., photostimulation) being reflected and the other one being transmitted so that the merged beams are co-aligned. Any incompatibilities can be resolved by inserting additional half-wave plates along the optical path. However, since no element has perfect transmission any additional optical element will reduce the overall power throughput. 3D SHOT is best assembled by first building the laser beam line at a fixed height on an optical table, then by installing the additional optical components starting from the beam merging cube or dichroic mirror, and in the sequential order outlined below.
-
1.
Fully assemble a standard multiphoton imaging microscope with the addition of a dichroic or polarizing beam splitter before the tube lens. Our experimental setup was built around a modified Movable Objective Microscope (MOM, Sutter Instrument Company), but alternate commercial and custom microscope designs may be used. We recommend using an additional 4-f relay of lenses (2-inch diameter, achromatic IR-coated doublets, 200 mm focal length), if the microscope’s image plane is not directly accessible at a location that is suitable to place the zero-order block.
-
2.
Place lens L4 and the SLM first so that the lens is at a distance f4 from the SLM and the image plane. If a beam expander is added along the laser path before the SLM, it is possible to use the experimental setup as a multiphoton holographic microscope. Several tests such as evaluating the accessible volume under the microscope objective, as well measuring the spatial dependence of losses in diffraction efficiency are possible and applicable in this form. However, in the final calibration procedure there will be a very fine evaluation of diffraction efficiency and accessible volume once all parts are included. By finely adjusting the position of lens L4 along the optical axis while the SLM displays a uniform phase mask, it is possible to precisely place the center of the 3D hologram in the center of the image, and at the same focal depth as the two-photon imaging plane.
The laser should operate at minimum power levels during the alignment procedure. Some laser models are fitted with co-aligned low-power red lasers that may be used to safely align the entire system with the exception of the diffraction grating tilt that must be aligned for the desired excitation wavelength. The SLM surface is one of the most sensitive areas and light should not be focused on it (this may happen when placing lens L3). The safest approach is to temporarily replace the SLM with a flat mirror and to align the SLM at the end by focusing undiffracted light on the zero-order block.
-
3.
The second design step is to replace the collimated beam illuminating the SLM with the engineered CTFP. For this, one must place and align lenses L3, L2, L1, in that order, in successive 4-f configurations. Pinholes and far-field images of the infrared beam can be used to check centering and beam collimation, respectively, to ensure that each newly inserted lens is properly centered and spaced. The reflective grating is placed last, at a distance f1 from lens L1 with its reflective surface orthogonal to the optical axis.
-
4.
The laser beam line must then be readjusted to illuminate the grating with an oblique incoming path so that the first diffracted order propagates back along the previously established optical axis. Placing temporary iris apertures on the lenses can help recentering the optical path. Fine adjustments of the beam propagation direction can be made by attaching the diffraction grating to a 2-axis tilt mount and precisely tuning the orientation of the diffracted beam.
-
5.
The final assembly step is to place the zero-order block at the focal spot of the default SLM image (uniform phase mask), where undiffracted light is expected to create a sharp focused spot, and the rotating diffuser at a distance f2 from lens L2. Various designs have been proposed to create a zero-order block and careful consideration should be given to choosing the adequate design. Most commercial SLMs are expected to allow a few percent of undiffracted laser light, which represents a significant amount of power on a small surface area. Thin metal films are not recommended because they cannot dissipate heat rapidly enough to avoid overheating. Instead, we suggest that one depolish the center of a flat optical surface (IR coated coverslip) with a small diamond-coated grinding tip. The depolished glass surface will diffuse most undiffracted light in all directions without accumulating heat. Alternatively, if a larger than necessary zero-order block is acceptable, a ~1 mm metal object, such as a neodymium magnet on a coverslip, will be able to withstand these powers.
During operation, special attention should be given to ensure that the rotating diffuser is spinning before allowing high laser power settings to avoid SLM damage.
2.2 Characterization and Performance Metrics for 3D-SHOT
Our primary goal is not to render a visually accurate hologram but instead to increase contrast for two-photon excitation at selected locations while avoiding inadvertent photoactivation of non-targeted areas, which relaxes constraints on hologram computation. Hence, the traditional metrics used to characterize imaging systems such as resolution, contrast, and speckle are not adequate to evaluate the capabilities of 3D-SHOT in experimental applications.
Instead, to evaluate the capabilities of 3D-SHOT and quantify how two-photon absorption is spatially distributed in 3D, we placed a thin fluorescent film on a microscope slide under the excitation objective to record the corresponding two-photon fluorescence image with a fixed sub-stage objective coupled to a camera (Fig. 2a). We recorded tomographic slice images by mechanically moving the excitation objective along the z axis by 1-μm increments. The resulting data correspond to a quantitative 3D measurement of two-photon absorption induced by the CTFP. We first consider the case of conventional 3D holography (Fig. 2b). We computed a 10 μm disk image target at z = 0 where we imposed a high-frequency speckle pattern to maximize spatial confinement along the z axis. Projection views of two-photon absorption along the y, x, and z axes show how, even in an optimized hologram, inadvertent photostimulation remains possible above and below a neuron targeted with this method. With 3D-SHOT however, experimental results (Fig. 2c) show that temporal focusing significantly enhances spatial confinement along the z axis in the CTFP.
Optical characterization of the spatial resolution of CGH vs 3D-SHOT. (a) We used a fluorescent calibration slide and an inverted microscope to quantify two-photon excitation in 3D. (b) For conventional holography, we consider a 10 μm diameter disk target, and show (from top to bottom) projection views of two-photon absorption in the (x,y), (z,y), and (z,x) planes. (c) With 3D-SHOT, the CTFP was adjusted to a 10 μm diameter target and the same projection views were recorded. (d) We measured the FWHM of the radial (top) or axial (bottom) PSF measured through brain slices of varying thickness (* indicates p < 0.05, Kruskal-Wallis Test with multiple comparison correction. Data represent the mean and standard deviation of n ≥ 5 observations for each thickness of brain tissue). (Adapted from Pegard et al. [7])
Since 3D-SHOT is developed for the primary-use case of optogenetic stimulation in brain tissue, we also recorded the effect of propagation through scattering medium on the radial and axial confinement of 3D-SHOT excitation. We cut acute mouse cortical brain slices of varying thickness and placed them between the excitation objective and the fluorescent slide. Recording two-photon absorption through physiologically relevant amounts of brain tissue revealed that scattering degraded radial resolution only after passing through 400 μm of mouse brain (Fig. 2d, 200 μm: p = 0.22, 300 μm: p = 0.21, 400 μm: p = 0.001, Kruskal-Wallis). Although the axial point spread function exhibited apparent degradation when imaged through brain tissue, this decrease was statistically significant only after passing through 300 μm of scattering tissue (Fig. 2d, 200 μm: p = 0.56, 300 μm: p = 0.04, 400 μm: p = 0.05, Kruskal-Wallis).
2.2.1 Scanless 2P Optogenetics Using 3D-SHOT
For neurobiological applications, we evaluated the spatial resolution of 3D-SHOT via quantitative measurements of the photocurrent amplitudes elicited by optogenetic stimulation in neurons. To do so, we expressed microbial opsins in neurons through in utero electroporation of mice embryos. Opsin expressing neurons were then brought under the objective either in acutely prepared cortical brain slices or in vivo with head-fixed animals.
To measure the spatial resolution of optogenetic excitation (or “Physiological” Point Spread Function – PPSF) we recorded the neuronal response to multiphoton photostimulation as a function of the displacement between the holographic target and the patched cell (Fig. 3a).
3D-SHOT generates axially confined photoactivation. (a) A photostimulation pattern generated with CGH (top) or 3D-SHOT (bottom) was mechanically stepped along the optical axis (z) and passed through a cell expressing opsin. Photocurrents were recorded in the whole-cell voltage-clamp configuration in neurons. (b) FWHM of the characteristic response profile for both methods at various power levels. (c) Photocurrent response profile for CGH (left) and 3D-SHOT (right) with a 10 μm disk target and different power levels. (d) Spatial profile of two-photon evoked spiking of a L2/3 pyramidal neuron in a mouse brain slice (left) in the radial dimension. Black: CGH; red: 3D-SHOT, p < 0.56 Mann-Whitney U-test, and (right), along the axial dimension (p < 0.006, Mann-Whitney U-test). (e) Quantification of the FWHM comparing CGH and 3D-SHOT. (f) Full volumetric assessment of photostimulation resolution, points throughout the volume were tested, but only points that elicited spike probability greater than zero are shown. (Adapted from Pegard et al. [7])
The efficacy of two-photon excitation is not only dependent on the shape of 3D-SHOT’s CTFP, but also on the precise targeting of this pattern to the cell soma, the level of opsin expression in the targeted neuron, and the laser intensity. Computer-generated holography already offers micron-level spatial resolution for placing holographic targets onto the desired neurons with a microscope objective. However, the level of opsin expression varies from neuron to neuron, and consequently so does the required power level for photostimulation. We experimentally compared (Fig. 3a) the spatial confinement of 3D-SHOT and 2P-CGH (without temporal focusing) for photoexcitation as a function of incident laser power density. Toward this end we obtained voltage-clamp recordings of neurons, and recorded the PPSF at a variety of different laser powers (Fig. 3b). With conventional holography, we observed substantial photocurrents 25–50 μm above and below the disk image target, indicating that photoactivation of non-targeted neurons is likely to be an issue. Temporal focusing significantly enhances spatial resolution with 3D-SHOT, and photocurrents are more significantly attenuated above and below the primary focus (Fig. 3c). We observed that the axial resolution with 3D-SHOT was significantly improved relative to CGH, even using several orders of magnitude more laser power. Whereas two-photon photoexcitation with CGH relies only on defocusing to confine the excitation light to the desired volume, 3D-SHOT benefits from simultaneous defocusing and temporal confinement, as femtosecond pulses are temporally stretched above and below the desired target which further attenuates the nonlinear response regardless of the targeted area in the (x,y) plane [41]. 3D-SHOT’s shallow relationship between laser power and spatial resolution is critical, in that it allows sufficient excitation light to generate action potentials without significant loss of spatial confinement, as it normally occurs with CGH, and gives the user the option to use additional power to reliably stimulate neurons when the exact level of opsin expression is unknown without significantly affecting spatial resolution.
2.2.2 3D-SHOT Photostimulation with Single-Neuron Resolution
We next quantified the physiological spatial resolution of CGH and 3D-SHOT in neurons by measuring the spiking probability along the radial direction in the imaging (x,y) plane and along the optical (z) axis. We compared holography and 3D-SHOT by projecting a single photostimulation target placed at a distance (x,y,z) from a patched neuron in mouse brain slice, either with single copy of the CTFP with 3D-SHOT or a disk-shaped pattern of equivalent size using CGH. We measured spike probability as the hologram was displaced in small increments by mechanically moving the objective relative to the patched cell. Experimental results (Fig. 3d) show similar spatial resolution with both methods in the radial direction in the focal plane, with a FWHM of 10 ± 2 μm for holography, and 9 μm± 1.3 for 3D-SHOT (p = 0.57, Mann-Whitney U-Test) consistent with the dimensions of the disc and Gaussian patterns at the focal plane (Fig. 4a).
3D-SHOT provides cellular resolution photostimulation in a large volume through digital focusing. (a) To quantify the spatial resolution of 3D-SHOT as a function of hologram target depth, we recorded the spike probability in cortical neurons while digitally targeting varying positions along the optical axis (z), and measuring resolution by mechanically sweeping the objective over the entire (z) range and measuring the response at each point. (b) Spike probability in cortical neurons while targeting the same cell from different axial displacements (p = 0.2, Kruskal-Wallis Test). (c) Spike probability resolution as a function of digital displacement – shaded green colors denote mechanical sweeps across the optical axis for different digital displacements. (d) Quantification of the FWHM for the axial fit of spike probability as a function of digital defocus from the focal plane (p = 0.17, Kruskal-Wallis Test). (Adapted from Pegard et al. [7])
However, with conventional holography, the spike probability along the z axis does not permit single-cell resolution (FWHM = 78 ± 6 μm). In contrast, 3D-SHOT provides far superior resolution (FWHM = 28 ± 0.7 μm, p = 0.006, Mann-Whitney U-Test, Fig. 3d) compatible with single-cell resolution in all three dimensions, in that the FWHM of spike probability is on par with the typical dimensions of a cortical neuron and their inter-somatic spacing (Fig. 3e). We recorded from neurons in brain slices and measured the spiking probability in response to 3D-SHOT excitation by digitally refocusing a hologram to stimulate positions throughout a 50 × 50 × 100 μm (x,y,z) grid. This experiment revealed that the neuron was photoactivated only when the disc image was targeted to the cell body (Fig. 3f).
2.2.3 Spatially Precise Remote Control with 3D-SHOT
A major advantage of holographic optogenetics is the ability to photoactivate neurons at different depths that are part of the same circuit. Since the major advance of 3D-SHOT is its ability to target temporally focused patterns arbitrarily in 3D, it is vital that 3D-SHOT maintains its ability to activate neurons with high spatial resolution even when digitally focusing light far from the zero-order of the optical system (e.g., the center of the optical axis at the natural focal depth of the system). Therefore, we next evaluated the accessible depth within the volume of interest by measuring the activation and spatial resolution as a function of the distance from the holographic zero order. Toward this end we measured spike probability in neurons via current clamp in mouse brain slices (Fig. 4a). To test if the CTFP can be digitally displaced along the z axis, we systematically moved the digital focus of the hologram (with a lens term on the SLM), and accordingly corrected the mechanical position of the objective by the same distance (δzDigital = −δzMechanical). This test showed that 3D-SHOT effectively photostimulates cells at locations distal to the zero order, as photocurrent and spike-probability were not affected by digital offset in z (Fig. 4b, p = 0.2, Kruskal-Wallis).
We next asked if the axial resolution of stimulation was constant when stimulating away from the natural focal plane at z = 0. For this we measured the FWHM of the axial PPSF as a function of digital defocus. As before, we digitally moved the holographic target along the z axis, but instead of matching the digital and mechanical offset, we stepped the objective across the entire range of the z axis and measured the physiological response at each location. This allowed us to measure the axial spatial resolution of photostimulation from locations distributed on either side of the optical axis. Results show that 3D-SHOT effectively confines excitation to the desired depth range throughout the 180 μm range that we sampled, as the FWHM of stimulation did not change as a function of digital defocus in z (Fig. 5c, d, p = 0.17, Kruskal-Wallis Test). These experiments show that 3D-SHOT retains axial confinement capabilities that are compatible with single-cell resolution for photocurrents and spike probability while targeting neurons at any depth within the accessible volume defined by the SLM and the microscope assembly.
Spatial resolution with simultaneous targets throughout a large volume. (a) 3D-SHOT was tested by simultaneously targeting 75 randomly distributed targets within the full operating volume defined by the SLM’s spatial range for the first diffracted order. Projection views of the 3D reconstruction of 2P-induced fluorescence in a calibration slide are shown along the (y, z), (x, z) axis, with a 3D projection. (b) Similar experiments were repeated with 20, 30, 50, and 75 targets. The FWHMs of the two-photon response were computed for each target, and show that spatial resolution and axial confinement are not significantly degraded by increasing the number of simultaneous targets in any given hologram (axial FWHM: p = 0.34, Kruskal-Wallis; radial FWHM: p < 0.001 for 75 ROIs; p > 0.05 for all other comparisons). (Adapted from Pegard et al. [7])
2.2.4 Volumetric Optogenetics at High Spatial Resolution
In addition to being able to create multi-target patterns at arbitrary depths with precise power control, the utility of 3D-SHOT for various applications is also determined by the absolute targetable volume and the number of targets that can be placed in a single hologram with the appropriate spatial resolution. To test the limits of the volume that can be simultaneously targeted using 3D-SHOT in our setup, we randomly selected up to 75 points distributed throughout the 350 μm × 350 μm × 280 μm volume of interest, and generated a point cloud hologram simultaneously targeting all of these points. We measured 2P absorption using a sub-stage camera (as in Fig. 2) and we measured the radial and axial confinement of each spot in the multi-target hologram (Fig. 5a). Quantifying the radial and axial FWHM showed that adding additional targets (up to 75) did not degrade the confinement of light in multi-spot holograms (Fig. 5b, p = 0.34 Axial FWHM, Kruskal-Wallis).
Overall, our experiments show that with 3D-SHOT as with CGH, the accessible volume and available optical power under the objective depends on the diffraction efficiency of the SLM, the laser power, and on cumulative losses across the optical path. Altogether, these design parameters determine the number of neurons that can be simultaneously illuminated with the desired spatial resolution. Here, with 600*800 pixels on the SLM, we characterized single-shot photostimulation of up to 75 targets (limited by laser power) with no degradation of resolution within a 0.034 mm3 volume (350 μm × 350 μm × 280 μm, Fig. 5). For comparison, custom photostimulation patterns have been demonstrated in previous works within a 0.017 mm3 operating volume with multi-level TF [51] (240 μm × 240 μm × 300 μm), ~6.25 × 10−4 mm3 with point scanning methods (250 μm × 250 μm × 15 μm) [18, 33].
2.3 Calibration of 3D-SHOT with Imaging System
The calibration and alignment of the optical system is critical to the successful use of any multiphoton stimulation system, this is made even more challenging when improving the resolution of such a system. Furthermore, whereas it is customary to report the best possible resolution in optics publications (to explain the potential of the technique), it is also known that the resolution is not constant throughout the entire working volume. However, for biological experiments, it is necessary to know what the actual resolution is at any given point in the working volume. Furthermore, even subtle errors introduced by aberrations of lenses and the SLM can lead to mistargeting problems that will prevent accurate experiments if they are not accounted for. To that end, we developed a calibration protocol and numeric tools to map the 3D holographic stimulation path with the multiplane two-photon imaging path. This calibration empirically accounts for aberration and deformities that are introduced both by the SLM and associated lenses, but also by the optomechanical defocusing method (in this example an electro tunable lens; ETL). This strategy provides both an improved calibration over less thorough procedures, and even more critically accurate measurements of the size of the holographic disk throughout the useable volume. While this procedure is quite slow, it is fully automated and can be run overnight (see Note 2.3.1). Scripts are available at https://github.com/adesniklab/3D-SHOT/AutoCalib
In addition to the 3D-SHOT and 2P imaging system described above, the calibration requires a substage camera (Fig. 6a). While many camera objective pairings are theoretically useable, we used a Basler camera (acA1300-200um) with a 5× Olympus air objective (Olympus MPLFLN) and a thin fluorescent slide (see Note 2.3.2). Care should be made to match the substage camera’s field of view to be at least as large as the imaging field of view.
Calibration protocol for 3D-SHOT. (a) Substage camera assembly for calibration with a uniform fluorescent thin film on a microscope slide. (b) A single hologram is imaged at 13 different planes by moving the hologram with respect to the thin fluorescent slide. The full range is ±40 μm from the estimated center of the hologram. (c) We fit a Gaussian curve to the measured fluorescence at each plane for each hologram recorded in (b). Relevant resolution characterization values (peak intensity, FWHM, and depth) are extracted for each hologram. (d) We first identify the relationship between the predicted SLM defocus and the detected depth of the corresponding holograms. (e) Mapped relationship between hologram FWHM and the hologram depth (left) is measured in the entire volume accessible by the SLM (right). (f) Hologram diffraction efficiency is measured throughout the field of view. (g) True depth of the two-photon imaging planes, as detected by the substage camera. We note that the planes are neither flat, evenly spaced, nor parallel to the axis of the camera, but that the calibration will account for all those discrepancies. (h) The final hole pattern in SLM space accounts for aberrations and curvature from both the SLM/stimulation path and imaging path. (i) Images of the holes ablated in the first plane, and for a subsequent plane. The hole pattern is asymmetric, so that subsequent planes will not burn in the same location. (j) Simulated targeting error over the full calibrated volume
Calibration Procedure
-
(a)
Manually position the substage camera such that the slide is in focus, and the illuminated area is in the center of the substage camera’s field of view. Tip: It may help to zoom in the 2P image and/or reduce the line count to get more visible fluorescence on the substage camera. However, care should be made not to photobleach the slide, and it will be necessary to return the imaging conditions to their normal settings before the rest of the calibration.
-
(b)
Compute 500–1000 test holograms containing a single target randomly placed throughout the imaging accessible volume. Tips: Random spots work slightly better than regularly placed spots to avoid overfitting.
-
(c)
Coarse Alignment. Sequentially illuminate each hologram with the same power and record the fluorescence on the substage camera (Fig. 6a). Move the objective mechanically in 25-μm steps increments throughout the useable volume to get a stack of images for each hologram. From this stack you can get the expected XY location of each hologram, and the peak fluorescence depth. Tips: Make sure that the power level you use here is neither saturating the substage camera, nor too dim to be visible. As holograms near the center of the optical axis will be brighter (better diffraction efficiency) we recommend using a test hologram near the zero order block and set the power such that it is just below saturating the camera pixels.
-
(d)
Fine Alignment. Take a z stack at 5–6 μm spacing for the ±40 μm around the expected Z location of each test hologram (Fig. 6b). Extract the fluorescence at the center of the hologram across the measured depths and fit this value to a Gaussian. The width of this Gaussian is the axial FWHM for the measured XYZ depth, and the peak of the Gaussian will be used to determine the diffraction efficiency for this point (Fig. 6c). The radial FWHM is determined by the fluorescence profile of the holographic spot at the best depth. It is best measured by fitting the observed fluorescence to a Gaussian, but as it is measured at a high spatial resolution already, this is not critical. Tip: while there may be aberrations or curvature from the SLM or other optical elements, they don’t need to be explicitly corrected as the general mapping strategy will account for all smooth distortions.
-
(e)
Fit SLM locations to substage cameras. For every SLM XYZ coordinate we now have a corresponding XYZ location measured by the substage camera with the depth determined from the z stack. Some holograms may need to be excluded if they were not properly detected (see Note 2.3.3). Use a polynomial fit to map SLMXYZ to CameraXYZ (Fig. 6d). Tip: When fitting make sure to scale both the camera pixels and the z depth to similar size units (such as μm) to prevent overweighting one axis. It is best practices to use ‘hold out’ data to test the fidelity of your fit. This will allow you to detect if there are systematic problems in your procedure.
-
(f)
At this point you will have collected all the necessary information to detect any variation in the shape of the holograms throughout the useable volume. Axial FWHM is often degraded when reaching the limits of your optical system (both radially near the edges, and axially at far defocus levels). It is important to restrict your experiments to locations where this FWHM is acceptable for your application (Fig. 6e).
-
(g)
To determine the diffraction efficiency as a function of target location in the accessible volume, you may divide the observed peak fluorescence from the fine calibration by the best fluorescence observed in the experiment. Here again, we recommend a polynomial interpolation to map the scalar “diffraction efficiency as a function of the SLMXYZ coordinates” (Fig. 6f). Tips: The diffraction efficiency accounts for many spatially dependent losses of observed fluorescence beyond holographic diffraction efficiency itself. This however works out to the more useful measure when running biological experiments. Furthermore, it would be more correct to get the best possible measurement of fluorescence with the zero-order block removed and a zero-order hologram. However, in practice this is unnecessary, the disruptions from removing and replacing the zero-order block are non-negligible, and the absolute value of the measured diffraction efficiency is less important than the exact profile along which it falls off.
-
(h)
Record the XYZ shape of each imaging plane. Due to aberrations induced by the ETL and the other lenses in the imaging path (and the substage camera itself), each imaging plane will have some three-dimensional shape, and they will be different for all defocusing levels. Image the slide at a single ETL-based depth and record the fluorescence on the substage camera. Then move the objective axially to create an orthoscopic image of its 3D profile. Repeat for many ETL defocus levels corresponding to the range you want to calibrate (Fig. 6g). Tips: Make sure to use enough power to be visible on the substage camera but not bleach. Increase the camera acquisition time, or frames averaged to get sufficient signal to noise.
-
(i)
Using a polynomial fit, map the measured Z depth to each ETL depth value as a function of XY position. Tips: you should be able to see the shape of each of your imaging planes, curvature and ripple in these planes is common and usually not detrimental to imaging quality or stimulation success.
-
(j)
To perform the final step in the alignment relating camera space to imaging space we calculate the SLM coordinates that are necessary to place holographic spots on each of the ETL planes that will be calculated. We use a grid of at least 16 targets per plane, each target offset laterally so that the projection of all targets onto a single plane will not be overlapping. Calculate single target holograms for each of these targets (Fig. 6h, i).
-
(k)
Ablate or bleach a hole in the fluorescent slide for each of this second set of holograms. Take a two-photon image of the slide at the appropriate ETL depth before and after each hole. The difference between this before and after image will reveal the XY position of each hole and the ETL depth can be used as the detected depth to create a 2PXYZ map for each of the second set of holograms. Tips: setting the ablation power can be tricky, higher peak energy, lower wattage pulses appear to bleach the slide more than they create cavitation and thus provide a more representative “hole”. We use 5x the standard imaging power for 500 ms to ablate a hole.
-
(l)
Fit the picked CameraXYZ locations to the detected 2PXYZ locations using a polynomial fit as above. The final calibration is 2PXY + ETLZ -> CameraXYZ -> SLMXYZ (Fig. 6j). Tips: All of these fits are symmetric so it can be self-checked by measuring the error by attempting to map coordinates onto themselves with a “round trip” interpolation between any two coordinate systems. Additionally, since the last step ablates targets for which both SLMXYZ and CameraXYZ coordinates are known, a second mapping can directly compare SLMXYZ to 2PXY + ETLZ coordinates, but since this approach does not include a true measurement of Z it is assumed to be less accurate. Typically, the mean interpolation error for a calibration is <2 μm throughout the entire useable area.
-
(m)
Troubleshooting. There are two very common sources of miscalibration. First, imaging conditions can change. This can occur due to evaporation of the immersion liquid, instability in lasers, or other sources. Sometimes the fluorescent intensity of holograms decreases over time. To detect this, plot the peak hologram intensity in order in which it was measured, as the locations are randomized, if any trend is visible it indicates some instability. Imprecise points can be manually excluded or the whole procedure can be repeated with steps taken to ensure that this problem will not occur again.
Second, the calibration slide may move. Since the calibration can take many hours, even subtle movement of the slide will disturb the calibration. This can be an insidious problem as depending on when this movement happens it can manifest in different ways. Prevention is the best remedy, and firmly securing the slide and the substage camera will mitigate this issue. A wise step at the end of a calibration is to conduct a hole test by ablating targets above and below a test location, with offsets of a few microns, to test the XZY accuracy (even 2 μm offset target will burn less efficiently than a properly targeted hologram). Typically, slide movements will only result in a XYZ offset and a digital offset can be applied without the need for a full recalibration. This approach can also help fix small post-calibration misalignments that may occur if there is any drift of the laser beam, when a full recalibration is not desired.
Notes
-
2.3.1: Overnight calibrations: While the described calibration is designed to run autonomously, several problems can arise that will disrupt it. First, the immersion liquid between the objective and the slide can dry up. To avoid this, we use a 1:10 dilution of Ultrasound Gel (NA 1.0, Parker Aquasonic 100), and create a well to hold excess liquid. Second, if the setup is much more susceptible to movements during calibration than it will be in its final form. We also recommend signage to ensure nobody disturbs the procedure.
-
2.3.2: Thin Fluorescent Slide: The detected resolution will be dependent on the thickness of the thin fluorescent slide. We manually spray Fluorescent Red (Tamiya TS-36) spray paint on standard microscope slides, and then screen each to be of uniform, minimal, and comparable thickness using the two-photon imaging system. As the diffraction-limited spot (DLS) of the imaging system is smaller than the 3D-SHOT spot, it is a useful benchmark for slide quality. For this step, we take a z-stack of the stationary imaging DLS using various slides. Variation of the same spot in the same location is caused by the slide thickness, and thus slides that report the smallest axial FWHM will be the thinnest. It is very important to pick a slide and a field of view that is even and flat. Small and sparse blemishes will be tolerated by the redundancy of the calibration, but large uneven fluorescence, or frequent scratches or holes will prevent accurate power calibrations.
-
2.3.3: There are many reasons why a given hologram may be poorly detected (e.g., out of range of the camera, insufficient diffraction efficiency, high noise), but these poorly read datapoints will increase the overall error of the calibration and decrease its usefulness. It is therefore better to have a smaller high-quality calibration than a larger one that contains inaccurate data. To ensure data quality, we set a brightness threshold for each hologram to be included in the calibration. The peak fluorescence should be at least 2× the Standard deviation of the imaging noise, points that do not satisfy this criterium are removed from the calibration data.
2.4 Comparison of Opsins for Precise Activation of Activity
Precisely timed neuronal activation cannot be achieved through technological progress in optical instrumentation alone. The molecular actuator, i.e., the opsin, is also a critical element for proper control of neural activity. The opsin must be selected deliberately, such that it synergizes with your chosen optical stimulation technique. In this section, we specifically discuss the selection of opsins that best leverage new capabilities introduced by 3D-SHOT to simultaneously illuminate the entire cell with strong photocurrents. Simultaneous illumination approaches as opposed to “spiral scanning” favor opsins with very fast kinetics and high peak amplitudes, enabling precise timing of action potentials with minimal jitter. Oppositely, asynchronous scanning-based approaches may benefit from slower opsins allowing for greater integration times, and can be activated with less laser power, but at the expense of temporal precision. Both approaches require opsins that are well-activated by 1030-nm light to best match commercially available high-power stimulation lasers.
The requirements for opsin constructs for 2-photon optogenetics differ considerably from those used for one-photon activation. Most notably, multiphoton approaches benefit from “soma-targeted opsin constructs”, i.e., those that are only expressed in the soma and proximal dendrites. Without soma targeting, the spatial resolution is compromised [11, 19, 56]. Furthermore, other opsin properties, such as photocurrent amplitude, absorption spectrum, and photocurrent kinetics, will strongly affect the experimental abilities of a 3D-SHOT system. These biophysical properties will interact with both the imaging and 3D-SHOT system and will alter the resolution, precision, and scale of neural control that is possible. We will briefly summarize how these properties interact, before describing a protocol for assessing opsin properties with regard to how to best activate or suppress a neuron. While many techniques are employed in this evaluation process, we will focus on those steps that are germane to opsin evaluation and two-photon optogenetics while directing the reader elsewhere for some technical procedures. While we will focus on selection of activating opsins, we will briefly discuss the additional concerns surrounding selection of a suppressing opsin.
We will also include, where possible and relevant, data on existing opsins. While many studies focus on individual features of opsin behavior, proper evaluation of an opsin requires a holistic understanding of many properties of those opsins. There are relatively few commonly used activating opsins used with multiphoton optogenetics the most common variants are ChR2 [28], C1V1 [8, 13], ChrimsonR, Chronos [15], ChroME [11], ReaChR [57], CoChR [56], and ChRmine [14] each with their own advantages and disadvantages. Opsins for two-photon suppression are less well characterized with only Arch [13], NpHR3, PsuACR, iC++, GtACR1 [11], and GtACR2 [58] being described.
We will select for opsins that:
-
Are well-trafficked to the somatic membrane with little toxicity.
-
Have large photocurrents.
-
Have fast kinetics.
-
Are well activated by 1030-nm light.
-
Are compatible with the all-optical approach of choice.
-
Can reliably drive spiking in vivo.
Procedure
-
1.
Subcellular Targeting.
When expressed in neurons, the opsin must be properly trafficked to the cell membrane but restricted to whatever extent possible away from the distal dendrites and axons (Fig. 7a). A sequence from the Kir2.0 channel [59] can be very helpful to export from endoplasmic reticulum, while a portion of the Kv2.1 channel [60] has become the standard (but not the only [56]) “soma targeting” motif, facilitating both membrane expression and restriction to the soma and proximal dendrites. It is advisable to use all subcellular targeting motifs even when testing opsins in reduced preparations such as CHO cells, as membrane targeting can affect photocurrents substantially. To begin opsin characterization, it is advisable to examine the trafficking of your opsin by creating a construct with the opsin fused to a fluorophore, even if ultimately a different fluorophore configuration is preferable. This way, one can ensure that the construct is well-trafficked to the membrane, while still restricted as much as possible to the soma. Furthermore, internal protein aggregation may be an indicator of toxicity. For more detailed discussion of toxicity assessment at multiple stages, see Note 2.4.1. In our experience, with the exception of incidences of overexpression, the gene delivery technique (e.g., AAV, IUE, transgenic) does not dramatically change intracellular trafficking patterns. Still, the most rigorous approach is to examine your particular opsin with the delivery mechanism you will use (see Note 2.4.2 for a discussion of delivery approaches).
Characterizing opsin characteristics for use with 3D-SHOT. (a) Comparison of non-soma-targeted opsin localization (left) to soma-targeted opsin localization (right). (Adapted from Mardinly et al. [11]). (b) Comparison of photocurrent FWHM (right) at different points on the opsin response function (left). Closer to saturation (dark blue), the actual FWHM of the photocurrent is larger than the theoretical opsin FWHM. (c) Comparison of photocurrent amplitude and kinetics for several commonly used opsins expressed in CHO cells. (Adapted from Sridharan et al. [61]). (d) Schematic of three common opsin kinetic metrics: left, time to peak used to measure opening kinetic. Center, desensitization. Right, tau off, a metric of decay kinetic. (e) Schematized absorption spectra for three opsin types compared with GCaMP absorption spectrum (dashed green). (f) Schematic of how, with fast imaging and slow opsins (top left) scan-induced photocurrents can accumulate to produce unwanted spiking, while in other conditions they decay and do not produce spiking. (g) Schematic displaying how under different stimulation conditions, short laser light pulses (left) can produce more or less reliable spike trains depending on opsin characteristics
-
2.
Photocurrents.
Photocurrent is perhaps the most obviously important measure of potency for an opsin. In many cases the opsin that fluxes the most current will be the most useful, as more potent opsins can drive more cells with less energy. While it may be convenient to compare the one-photon response to light, 3D-SHOT relies on the multiphoton process and thus one-photon responses are not an acceptable substitute (see Note 2.4.3).
There are two main criteria to consider when evaluating photocurrent. First, it is important to probe a large range of light intensities. Different opsins will saturate (i.e., reach maximal photocurrent) at different illumination powers and at different photocurrent levels. While it is necessary to reach a certain threshold to spike a cell (1 nA is a good approximation to spike a L2/3 pyramidal cell with a 5-ms pulse), the shape of this curve will impact your resolution. The minimal power needed to spike a cell will dictate the total number of cells that can be activated with a given microscope, and the total heat added for a given number of activated cells (see Note 2.4.4). In addition, the further this current is from the saturating current the better the effective resolution will be (Fig. 7b).
While peak current is often reported, the average current over a given pulse duration, or the total charge fluxed, is what ultimately drives cell activation and thus is a more relevant measure for photoactivation. This is especially important considering that many opsins show desensitization (see detailed discussion in step 3, Opsin Kinetics).
Available opsins differ greatly in photocurrent magnitude (Fig. 7c). While direct comparisons of all commonly used opsins is not available (though see Sridharan et al. [61] for comparison of many), only ChroME family [11, 61] and ChRmine family [14, 62] opsins appear to reliably reach the 1 nA benchmark. Cells expressing either CoChR [56], and ReaChR [57] occasionally reached this threshold, but not reliably. It is possible that further improvement of targeting and expression will help these opsins reach this benchmark.
-
3.
Opsin Kinetics.
While selecting the opsin with the maximum photocurrent makes sense in many cases, it may come at the expense of speed. Fast opsins are necessary to take full advantage of the temporal control offered by 3D-SHOT and to write specific spike trains, with low spike jitter and high fidelity.
A fast opsin must both open and close its ion channel very quickly. If the closing kinetic is too slow, two or more action potentials may result from a single stimulation [34, 52, 53], and it may be impossible to drive cells to fire at high rates, disrupting the ability to write a known train. Similarly if the opsin opening kinetic is too slow, longer stimulation periods will be needed to drive the cell to fire and the uncertainty (jitter) in action potential timing [52] will increase.
Moreover, the opening (but not closing) kinetics of both activating and suppressing opsins [11, 46, 63] are often dependent on laser stimulation power, adding additional complexity [11]. As the precise mechanics that lead the opsin to transition between conducting states is not yet fully understood [64, 65], and even less is known about how these transitions may be affected by the two-photon process, there is often no way to model or infer kinetics based on the structure of an opsin alone. Instead, the only solution is to measure these kinetics with each new prospective opsin.
The opening kinetic is often measured using the time to peak or 90% current, but τ derived from an exponential fit can also be reported [8, 11, 15, 56] (Fig. 7d). During prolonged pulses, many opsins’ photocurrents reduce with time, a phenomenon known as desensitization (Fig. 7d). This may be either through inactivation of a population of channels and/or by individual channels entering states with different conductance [53, 65]. At the cessation of a pulse, the opsin current decays gradually. This closing kinetic is typically reported by fitting an exponential fit and calculating the τ (Fig. 7d) [56]. At times, a double exponential may better fit the data than a single one [11].
Several groups have endeavored to identify or engineer very fast opsins for one- or two-photon use [11, 15, 66,67,68,69]; these promise to be valuable tools for optogenetic control. While mutations that speed up an opsin often come at the expense of total photocurrent, this does not appear to be an absolute rule (note the success of ChroME and ChETA [11, 68]). Furthermore, it is hard to know how fast is “fast enough”? Chronos and the mutant forms, ChroME opsins, are the fastest opsins used in 2p-activation to date, and can be used in vivo to drive spike trains with jitter much less than one millisecond [11, 61, 70]. Nevertheless, in the correct conditions even much slower opsins such as ChrimsonR [52], CoChR [56], or ChRmine [14] can reach jitter of about 1 ms. However, cells expressing these last three opsins struggle to follow rates over 20 Hz [14, 52], probably due to their slower off kinetics.
-
4.
Two-Photon Spectra.
The two-photon absorption spectrum of an opsin affects its compatibility with imaging approaches and suitability for use with the high-power lasers typically used for 3D-SHOT. A chief concern with simultaneous imaging and holographic activation is the phenomenon of crosstalk, in which the scanning diffraction-limited spot used to excite GCaMP fluorescence also activates opsin molecules (the reader may also refer to Chaps. 2, 3, 5, 6, and 11 of this book for an extended overview of this phenomenon). Opsins with low absorption in the range of wavelengths used to image GCaMP (typically 910–930 nm) reduce crosstalk. Unfortunately, many traditional opsins are highly activated by blue light, so this has required the development of many new opsins. Alternate calcium indicators that absorb in other wavelengths are also available but are much dimmer than available GCaMPs [71,72,73]. In addition, most commercially available high-energy lasers emit around 1030 nm [63]. Thus, the optimal opsin would have a peak photocurrent around 1030 nm with a comparative minimum at the wavelengths to image GCaMP (~930 nm) (Fig. 7e, see Note 2.4.5 for further discussion of alternate strategies).
It is well known that fluorophores have two-photon absorption spectra that are considerably different from their one-photon counterparts [74]. To assess the sensitivity at various 2 photon wavelengths, the simplest approach is to use a 2p imaging microscope with femtosecond laser (e.g., Ti: Sapphire oscillator) that is tunable across a large range of wavelengths. Since spectral response is not known to be affected by the light delivery method, using a scanning imaging system to photoactivate opsins is a suitable approach to compare the relative activation at different wavelengths and measure the spectral response profile. Recording photocurrents from CHO cells at different wavelengths while scanning will provide a two-photon spectrum for the opsin. Emission power varies with wavelength, so be sure to test power out of the objective at all testing wavelengths.
Of the common excitatory opsins, ChR2 [28] and CoChR [56] are blue-shifted making them suboptimal for pairing with GCaMP imaging (Fig. 7e). Several 2p-optimized opsins, including Chronos, ChroME [11], and ReachR [57] peak around 1000 nm, and more red-shifted opsins such as C1V1 [13], ChrimsonR [11], and ChRmine [14] peak beyond 1040 nm.
-
5.
Characterizing Crosstalk in Imaging Conditions.
In most experiments, multiphoton optogenetics will be paired with multiphoton imaging. It is important to expressly consider the relevant ways that these two systems interact. While the stimulation laser can create an artifact on the imaging system (see Note 2.4.6), the more insidious form of crosstalk is where the imaging laser can activate the opsin. This crosstalk results from opsins that are somewhat sensitive to imaging wavelengths, as discussed above. Even when these depolarizations (or hyperpolarization in the case of inhibitory opsin) are not enough to overtly change the spiking rate of cells they can cause significant currents which may alter the timing of action potentials.
Opsin kinetics and strength also influence compatibility with 2p imaging. A diffraction-limited spot is used for imaging scans across a cell in a very different way than 3D-SHOT. Here, fast opsins are again advantageous because activation will decay to baseline between imaging frames, thus not driving the cell to spike (Fig. 7f). Strong, slow opsins may be most vulnerable to the effects of crosstalk, but this can be addressed at least in part by interleaving several frames in different areas to extend the time between repeated stimulation. In contrast, as activation is proportional to dwell time of the imaging laser on a cell, approaches that increase this dwell time, such as having a smaller field of view, will suffer worse crosstalk [11].
Due to the many variables affecting crosstalk currents, including imaging speed, power, wavelength, opsin kinetics, and more, it is important to test the actual currents induced in your typical imaging preparation [11]. Empirical measurements with your opsin of interest and in the typical imaging conditions are essential to understand the level of crosstalk that you will experience. Whole-cell recordings, even in ex vivo slice, under analogous imaging conditions will give the resolution needed to observe how large the imaging-induced photocurrents are.
-
6.
Characterizing In Vivo Spike Fidelity.
Ultimately the end goal of selecting an opsin is to cause neurons to fire action potentials. While many opsin/stimulation combinations can drive cell spiking [11, 34, 52] it is important to quantify the fidelity of this spiking. One should quantify to the fraction of cells spikeable, the fidelity of response (i.e., the fraction of pulses in which the cell spikes), the reproducibility of a response (i.e., the fraction of pulses resulting in one and only one spike, also known as no “doublets”), and the jitter of the resulting action potentials (to understand the limits of your timing control).
Furthermore, these evaluations should be performed at a variety of frequencies, and in conditions most similar to your biological experiments possible. Under different stimulation patterns and strengths, opsins may be more or less faithful (Fig. 7g), such as strong, slow opsins producing doublets at high stimulation powers. We recommend cell-attached or whole-cell recordings from the anesthetized animal. Of these in vivo criteria, being able to reliably evoke one and only one action potential per pulse is one of the most challenging and useful features. While this analysis has not been performed on all opsins both ChroME [11] and ChrimsonR [52] can be driven in the regime where one and only one spike is possible.
-
7.
Considerations for Multiphoton Suppression.
Multiphoton suppression involves using an inhibitory or suppressive opsin (one that hyperpolarizes a cell) instead of an activating one to prevent a cell from firing. Multiphoton suppression employs many of the same concerns as multiphoton activation. Concerns around spectrum, photocurrents, imaging compatibility, and toxicity requirements are all very similar between activating and suppressing opsins.
The primary difference is in the requirements for kinetics, whereas activation requires a fast off kinetic to prevent doublets; suppressing opsins have no such constraints. In fact, slower off kinetics can be helpful, as they allow discontinuous (aka duty cycled) light to achieve continuous inhibition. Similarly, as it is rare to know the precise timing of a naturally occurring spike, there is a diminished need for a fast on kinetic, but this comes at the cost of needing to suppress activity for a long duration. With increased photostimulation durations come increased hazards of heat buildup, and corruption from optical artifacts. Furthermore, opsins that desensitize to two-photon stimulation, i.e., have a high peak current but a lower sustained current, such as iC++ [11], are disadvantaged as the peak current contributes less to the overall suppression of activity.
Finally, whereas photoactivation is a nearly binary process, a spike is either evoked or it is not, suppression is graded. Even if a given opsin stimulation can suppress spontaneous activity, a particularly large endogenous stimulus might overwhelm this inhibition. As such many aspects of benchmarking inhibitory opsins are harder. Nonetheless, GtACR2 [58], GtACR1, and Arch [11] have all been used to successfully suppress activity in vivo. Although of these Arch and GtACR2 only suppressed ~50% of spontaneous activity [11, 58], whereas GtACR1 was more effective [11].
3 Conclusion
When selecting an activating opsin for very precise manipulation of spike trains there are a variety of factors that need to be weighed. Thus, a holistic approach that evaluates all qualities of the opsin is required to pick the optimal tool. Of the opsins that have been tested for their compatibility with two-photon optogenetics, only ChroME, ChRmine, CoChR, and ReaChR reach the photocurrent benchmark of 1 nA. Of these only ChroME, and ChRmine can be used to drive spikes with sub-millisecond jitter, and only ChroME responded reliably at rates above 40 Hz. However, as more opsins are developed for multiphoton use, this set of useable opsins will continue to grow. In the near future, there will be a generation of new opsins that are discovered or mutated from existing opsins that will advance our ability to control the firing of cells. As the field grows out of its infancy it is likely that more criteria will present themselves as essential for selecting the best opsin. Until they do, we hope this guide will aid in the benchmarking of future opsins for the precise recreation of neuronal activity patterns.
Notes
-
2.4.1: While observing aggregates in structural imaging is a clear indicator of possible toxicity, many exogenous proteins can have deleterious effects on the health of cells without necessarily forming aggregates. Thus, we recommend further assessment of cell health for all preparations used for experiments. Viral overexpression and/or combination with other stressors, including calcium buffering from GCaMP, may further lead to cells no longer responding physiologically. It is important to benchmark the health of cells with your chosen opsin and expression strategy. There are two broad categories to examine: First the intrinsic properties of cells, and second their physiological responses to stimuli. Intrinsic properties, such as a cell’s input resistance, resting membrane potential, membrane time constant, action potential threshold, and shape of an action potential should all remain unchanged, between opsin expressing and opsin negative cells, and are easily measurable in ex vivo whole-cell recordings. Similarly, the resting firing rate of cells in vivo should not be altered by the presence of an opsin, nor when imaging with standard GCaMP imaging conditions. Physiological assessment of cell health is more challenging and is often overlooked. Ideally, one would measure the in vivo responses to given sensory stimuli and show that they do not change with presence or activation of the opsin.
-
2.4.2: Opsin expression. While AAV delivery of opsin is a popular and effective strategy for expression in neurons, it can introduce variability, especially dependent on differences in the viral preparation [75]. Especially if new constructs are being created, the time to develop new viral delivery systems may be preventative. For this reason, we recommend using transfected CHO cells for opsin biophysics such as kinetics, and in utero electroporated cortical neurons for experiments assessing neuronal responses. For photocurrent assessment, while CHO cells will provide some information, we recommend using neurons. Subcellular targeting and the interactions between endogenous channels and opsins could potentially alter the effectiveness of different opsin constructs. For this reason, we recommend whole-cell voltage-clamp recordings in neurons in utero electroporated with the opsin construct of interest.
-
2.4.3: The two-photon response of opsins: while relatively little work has been devoted to comparing the two-photon responses of opsins, several lines of evidence support that the activation we see is indeed a two-photon process. First, others [13, 46] have fit very low power activation of C1V1 or ChR2 and concluded that it better followed a quadratic fit rather than a linear one. More recent work has shown [16] that a dispersed laser pulse, with lower peak energy, is unable to activate ChrimsonR while a compressed pulse is.
-
2.4.4: The risk of brain heating is greatly expanded with multiphoton imaging and optogenetics. It has been shown that even modest temperature increases used in one photon optogenetics can sometimes affect firing rates of cells [76, 77]. Thus, the much higher energies used in multiphoton optogenetics, and to a lesser extent imaging, could in theory lead to aberrant behavior. This concern is mitigated somewhat by a 2–3 °C decrease in the overall temperature of the brain simply through the cranial window and water immersion objective that is used in the microscope [78, 79]. Nonetheless, brain heating especially with multiphoton optogenetics remains a risk [80, 81].
-
2.4.5: While, by far the majority of groups doing all-optical multiphoton optogenetics use red-shifted opsins [11, 14, 16, 34, 82], others have focused on blue-shifted opsins with red indicators [58]. In theory, 3D-SHOT should be fully compatible with a lower wavelength laser. However, 3D-SHOT requires a high-energy laser to stimulate many cells. As currently there are few available lasers that have very high peak power at 930 nm, it is difficult to build a system that can control many cells. Nonetheless, this approach offers several advantages that should not be overlooked. All opsins recorded to date have substantial “blue shoulders”, i.e., current fluxed at lower wavelengths than their peak. As such with a red-shifted opsin there will be some activation by the imaging laser. This “scan activation” presents a potential confound that if not accounted for could change the results of a biological experiment.
-
2.4.6: Stimulation laser-induced artifacts in imaging. Just as the imaging laser can somewhat activate the opsin, so too can the 1030 nm stimulation laser drive GCaMP fluorescence, albeit sub-optimally. This creates an artifact whereby pixels recorded during stimulation will be contaminated and will appear brighter, in severe cases (such as when stimulating many targets) this artifact can be much brighter than the GCaMP fluorescence. Some groups have opted to exclude frames or pixels containing artifact [14, 18] but this can be preventative when stimulating at high rates, or for long durations, as is required during optogenetic suppression. Instead, we recommend syncing the firing of the stimulation laser to the phase of the fast-resonant mirror. By only allowing the stimulation laser to fire during the edge or flyback of the imaging field of view we ensure that very few neurons are obscured by the stimulation artifact. This can be achieved via a fast analog or digital circuit that controls a sufficiently fast electro-optic modulator controlling the stimulation laser’s power. As the rate of the resonant mirror is ~8 kHz, this modulated cycle will be approximately ~16 kHz much faster than any known opsins on or off rate. This very fast duty cycle ends up being a significant advantage. While this “gating” of the laser might exclude up to 50% of the total energy hitting the sample, we see photocurrents that are only reduced by 15–20%.
References
Adam Y et al (2019) Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature. https://doi.org/10.1038/s41586-019-1166-7
Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V (2014) Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron 84:1157–1169
Carrillo-Reid L, Han S, Yang W, Akrouh A, Yuste R (2019) Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell 178:447–457.e5
Sohal VS, Zhang F, Yizhar O, Deisseroth K (2009) Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459:698–702
Liu X et al (2012) Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. https://doi.org/10.1038/nature11028
Naka A et al (2019) Complementary networks of cortical somatostatin interneurons enforce layer specific control. elife. https://doi.org/10.7554/eLife.43696
Pégard NC et al (2017) Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nat Commun. https://doi.org/10.1038/s41467-017-01031-3
Packer AM et al (2012) Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods 9:1202–1205
Nikolenko V, Poskanzer KE, Yuste R (2007) Two-photon photostimulation and imaging of neural circuits. Nat Methods. https://doi.org/10.1038/nmeth1105
Ronzitti E, Emiliani V, Papagiakoumou E (2018) Methods for three-dimensional all-optical manipulation of neural circuits. Front Cell Neurosci. https://doi.org/10.3389/fncel.2018.00469
Mardinly AR et al (2018) Precise multimodal optical control of neural ensemble activity. Nat Neurosci. https://doi.org/10.1038/s41593-018-0139-8
Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY (2013) ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci 16:1499–1508
Prakash R et al (2012) Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods 9:1171–1179
Marshel JH et al (2019) Cortical layer-specific critical dynamics triggering perception. Science (80- ). https://doi.org/10.1126/science.aaw5202
Klapoetke NC et al (2014) Independent optical excitation of distinct neural populations. Nat Methods 11:338–346
Gill JV et al (2020) Precise holographic manipulation of olfactory circuits reveals coding features determining perceptual detection. Neuron 1–12. https://doi.org/10.1016/j.neuron.2020.07.034
Dalgleish HWP et al (2020) How many neurons are sufficient for perception of cortical activity? elife 9:1–99
Carrillo-Reid L, Yang W, Bando Y, Peterka DS, Yuste R (2016) Imprinting and recalling cortical ensembles. Science (80- ). https://doi.org/10.1126/science.aaf7560
Chettih SN, Harvey CD (2019) Single-neuron perturbations reveal feature-specific competition in V1. Nature 567:334–340
Rickgauer JP, Deisseroth K, Tank DW (2014) Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci 17:1816–1824
Daie K, Svoboda K, Druckmann S (2021) Targeted photostimulation uncovers circuit motifs supporting short-term memory. Nat Neurosci 24:259–265
Clancy KB, Schnepel P, Rao AT, Feldman DE (2015) Structure of a single whisker representation in layer 2 of mouse somatosensory. Cortex 35:3946–3958
Sato TR, Gray NW, Mainen ZF, Svoboda K (2007) The functional microarchitecture of the mouse barrel cortex. PLoS Biol 5:1440–1452
Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC (2005) Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433:597–603
Svoboda K, Yasuda R (2006) Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron 50:823–839
Svoboda K, Denk W, Kleinfeld D, Tank DW (1997) In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature 385:161–165
Denk W, Strickler JH, Webb WW (1990) Two-photon laser scanning fluorescence microscopy. Science (80- ). https://doi.org/10.1126/science.2321027
Rickgauer JP, Tank DW (2009) Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A 106:15025–15030
Katona G et al (2012) Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Methods 9:201–208
Reddy GD, Kelleher K, Fink R, Saggau P (2008) Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci 11:713–720
Yang W et al (2016) Simultaneous multi-plane imaging of neural circuits. Neuron. https://doi.org/10.1016/j.neuron.2015.12.012
Piyawattanametha W et al (2006) Fast-scanning two-photon fluorescence imaging based on a microelectromechanical systems two- dimensional scanning mirror. Opt Lett 31:2018–2020
Packer AM, Russell LE, Dalgleish HWP, Häusser M (2014) Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods 12:140
Yang W, Carrillo-Reid L, Bando Y, Peterka DS, Yuste R (2018) Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions. elife. https://doi.org/10.7554/eLife.32671
Zhang J, Pégard N, Zhong J, Adesnik H, Waller L (2017) 3D computer-generated holography by non-convex optimization. Optica. https://doi.org/10.1364/optica.4.001306
Eybposh M, Caira N, Atisa M, Chakravarthula P, Pegard N (2020) DeepCGH: 3D computer-generated holography using deep learning. Opt Express. https://doi.org/10.1364/oe.399624
Gerchberg RW, Saxton WO (1972) A practical algorithm for the determination of phase from image and diffraction plane pictures. Optik (Stuttg) 35:237–246
Nikolenko V et al (2008) SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators. Front Neural Circuits 2:5
Papagiakoumou E et al (2010) Scanless two-photon excitation of channelrhodopsin-2. Nat Methods 7:848
Tal E, Oron D, Silberberg Y (2005) Improved depth resolution in video-rate line-scanning multiphoton microscopy using temporal focusing. Opt Lett 30:1686–1688
Zhu G, van Howe J, Durst M, Zipfel W, Xu C (2005) Simultaneous spatial and temporal focusing of femtosecond pulses. Opt Express 13:2153–2159
Spesyvtsev R, Rendall HA, Dholakia K (2015) Wide-field three-dimensional optical imaging using temporal focusing for holographically trapped microparticles. Opt Lett 40:4847–4850
Durst ME, Zhu G, Xu C (2006) Simultaneous spatial and temporal focusing for axial scanning. Opt Express 14:12243–12254
Mayblum T, Schejter A, Dana H, Shoham S et al (2015) SPIE BiOS 932928
Prevedel R et al (2016) Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat Methods 13(12):1021–1028
Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A (2010) Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci U S A 107:11981–11986
McCabe DJ et al (2011) Spatio-temporal focusing of an ultrafast pulse through a multiply scattering medium. Nat Commun 2:447
Dana H, Shoham S (2011) Numerical evaluation of temporal focusing characteristics in transparent and scattering media. Opt Express 19:4937–4948
Therrien OD, Aubé B, Pagès S, De Koninck P, Côté D (2011) Wide-field multiphoton imaging of cellular dynamics in thick tissue by temporal focusing and patterned illumination. Biomed Opt Express 2:696–704
Bègue A et al (2013) Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation. Biomed Opt Express 4:2869–2879
Hernandez O et al (2016) Three-dimensional spatiotemporal focusing of holographic patterns. Nat Commun 7:1–10
Chen IW et al (2019) In vivo submillisecond two-photon optogenetics with temporally focused patterned light. J Neurosci. https://doi.org/10.1523/JNEUROSCI.1785-18.2018
Fenno L, Yizhar O, Deisseroth K (2011) The development and application of optogenetics. Annu Rev Neurosci 34:389–412
Gerchberg RW, Saxton WO (1972) Phase retrieval by iterated projections. Optik (Stuttg) 35:237
Sinclair G et al (2004) Interactive application in holographic optical tweezers of a multi-plane Gerchberg-Saxton algorithm for three-dimensional light shaping. Opt Express 12:1665–1670
Shemesh OA et al (2017) Temporally precise single-cell-resolution optogenetics. Nat Neurosci 20:1796–1806
Chaigneau E et al (2016) Two-photon holographic stimulation of ReaChR. Front Cell Neurosci. https://doi.org/10.3389/fncel.2016.00234
Forli A et al (2018) Two-photon bidirectional control and imaging of neuronal excitability with high spatial resolution in vivo. Cell Rep. https://doi.org/10.1016/j.celrep.2018.02.063
Stockklausner C, Ludwig J, Ruppersberg JP, Klöcker N (2001) A sequence motif responsible for ER export and surface expression of Kir2.0 inward rectifier K+ channels. FEBS Lett. https://doi.org/10.1016/S0014-5793(01)02286-4
Lim ST, Antonucci DE, Scannevin RH, Trimmer JS (2000) A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron 25:385–397
Sridharan S et al (2022) High-performance microbial opsins for spatially and temporally precise perturbations of large neuronal networks. Neuron 110(7):1139–1155.e6
Kishi KE et al (2022) Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185:672–689.e23
Ronzitti E et al (2017) Submillisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of chronos. J Neurosci. https://doi.org/10.1523/JNEUROSCI.1246-17.2017
Schneider F, Grimm C, Hegemann P (2015) Biophysics of channelrhodopsin. Annu Rev Biophys. https://doi.org/10.1146/annurev-biophys-060414-034014
Kuhne J et al (2019) Unifying photocycle model for light adaptation and temporal evolution of cation conductance in channelrhodopsin-2. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1818707116
Berndt A et al (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1017210108
Kato HE et al (2018) Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature. https://doi.org/10.1038/s41586-018-0504-5
Gunaydin LA et al (2010) Ultrafast optogenetic control. Nat Neurosci. https://doi.org/10.1038/nn.2495
Rajasethupathy P et al (2015) Projections from neocortex mediate top-down control of memory retrieval. Nature. https://doi.org/10.1038/nature15389
Ronzitti E, Conti R, Zampini V, Tanese D, Foust AJ, Klapoetke N, Boyden ES, Papagiakoumou EV (2017) Sub-millisecond optogenetic control of neuronal firing by two-photon holographic photoactivation of Chronos. J Neurosci 37:1246–17. https://doi.org/10.1101/062182
Dana H et al (2016) Sensitive red protein calcium indicators for imaging neural activity. elife 5:e12727
Hussein W, Berlin S (2020) Red photoactivatable genetic optical-indicators. Front Cell Neurosci 14:113
Mohr MA et al (2020) jYCaMP: an optimized calcium indicator for two-photon imaging at fiber laser wavelengths. Nat Methods. https://doi.org/10.1038/s41592-020-0835-7
Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A (2011) Two-photon absorption properties of fluorescent proteins. Nat Methods. https://doi.org/10.1038/nmeth.1596
Allen BD, Singer AC, Boyden ES (2015) Principles of designing interpretable optogenetic behavior experiments. Learn Mem. https://doi.org/10.1101/lm.038026.114
Owen SF, Liu MH, Kreitzer AC (2019) Thermal constraints on in vivo optogenetic manipulations. Nat Neurosci. https://doi.org/10.1038/s41593-019-0422-3
Moser E, Mathiesen I, Andersen P (1993) Association between brain temperature and dentate field potentials in exploring and swimming rats. Science (80- ). https://doi.org/10.1126/science.8446900
Podgorski K, Ranganathan G (2016) Brain heating induced by near-infrared lasers during multiphoton microscopy. J Neurophysiol. https://doi.org/10.1152/jn.00275.2016
Roche M et al (2019) In vivo imaging with a water immersion objective affects brain temperature, blood flow and oxygenation. elife. https://doi.org/10.7554/eLife.47324
Schmidt E, Oheim M (2020) Infrared excitation induces heating and calcium microdomain hyperactivity in cortical astrocytes. Biophys J. https://doi.org/10.1016/j.bpj.2020.10.027
Picot A et al (2018) Temperature rise under two-photon optogenetic brain stimulation. Cell Rep. https://doi.org/10.1016/j.celrep.2018.06.119
Zhang Z, Russell LE, Packer AM, Gauld OM, Häusser M (2018) Closed-loop all-optical interrogation of neural circuits in vivo. Nat Methods. https://doi.org/10.1038/s41592-018-0183-z
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2023 The Author(s)
About this protocol
Cite this protocol
Oldenburg, I.A., Bounds, H.A., Pégard, N.C. (2023). High-Speed All-Optical Neural Interfaces with 3D Temporally Focused Holography. In: Papagiakoumou, E. (eds) All-Optical Methods to Study Neuronal Function. Neuromethods, vol 191. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2764-8_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2764-8_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2763-1
Online ISBN: 978-1-0716-2764-8
eBook Packages: Springer Protocols