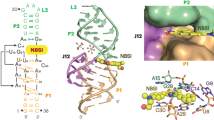
Abstract
Fluorogenic RNA aptamers are synthetic RNAs that have been evolved by in vitro selection methods to bind and light up conditionally fluorescent organic ligands. Compared with other probes for RNA detection, they are less invasive than hybridization-based methods (FISH, molecular beacons) and are considerably smaller than fluorescent protein-recruiting systems (MS2, Pumilio variants). Fluorogenic aptamers have therefore found widespread use as genetically encodable tags for RNA detection in live cells and have also been used in combination with riboswitches to construct versatile metabolite sensors for in vitro use. Their success builds on a fundamental understanding of their three-dimensional structure to explain the mechanisms of ligand interaction and to rationally design functional aptamer devices. In this protocol, we describe a supramolecular FRET-based structure probing method for fluorogenic aptamers that exploits distance- and orientation-dependent energy transfer efficiencies between site-specifically incorporated fluorescent nucleoside analogs and non-covalently bound ligands, exemplified by 4-cyanoindol riboside (4CI) and the DMHBI+-binding RNA aptamer Chili. This method yields structural restraints that bridge the gap between traditional low-resolution secondary structure probing methods and more elaborate high-resolution methods such as X-ray crystallography and NMR spectroscopy.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Weeks KM (2010) Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 20:295–304
Ge P, Zhang S (2015) Computational analysis of RNA structures with chemical probing data. Methods 79–80:60–66
Tian S, Das R (2016) RNA structure through multidimensional chemical mapping. Q Rev Biophys 49:e7
Micura R, Höbartner C (2020) Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem Soc Rev 49:7331–7353
Perez-Gonzalez C, Lafontaine DA, Penedo JC (2016) Fluorescence-based strategies to investigate the structure and dynamics of aptamer-ligand complexes. Front Chem 4:33
Wilhelmsson LM (2010) Fluorescent nucleic acid base analogues. Q Rev Biophys 43:159–183
Souliere MF, Haller A, Rieder R et al (2011) A powerful approach for the selection of 2-aminopurine substitution sites to investigate RNA folding. J Am Chem Soc 133:16161–16167
Souliere MF, Micura R (2014) Use of SHAPE to select 2AP substitution sites for RNA-ligand interactions and dynamics studies. Methods Mol Biol 1103:227–239
Stivers JT (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res 26:3837–3844
Haller A, Souliere MF, Micura R (2011) The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res 44:1339–1348
Katilius E, Katiliene Z, Woodbury NW (2006) Signaling aptamers created using fluorescent nucleotide analogues. Anal Chem 78:6484–6489
Gustmann H, Segler AJ, Gophane DB et al (2019) Structure guided fluorescence labeling reveals a two-step binding mechanism of neomycin to its RNA aptamer. Nucleic Acids Res 47:15–28
Xie Y, Dix AV, Tor Y (2009) FRET enabled real time detection of RNA-small molecule binding. J Am Chem Soc 131:17605–17614
Sinkeldam RW, Tor Y (2019) FRET assay for ligands targeting the bacterial A-site RNA. Methods Mol Biol 1973:251–260
Rieder R, Lang K, Graber D et al (2007) Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem 8:896–902
Haller A, Rieder U, Aigner M et al (2011) Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7:393
Steinmetzger C, Palanisamy N, Gore KR et al (2019) A multicolor large stokes shift fluorogen-activating RNA aptamer with cationic chromophores. Chemistry 25:1931–1935
Steinmetzger C, Bessi I, Lenz AK et al (2019) Structure-fluorescence activation relationships of a large Stokes shift fluorogenic RNA aptamer. Nucleic Acids Res 47:11538–11550
Passow KT, Harki DA (2018) 4-Cyanoindole-2′-deoxyribonucleoside (4CIN): a universal fluorescent nucleoside analogue. Org Lett 20:4310–4313
Steinmetzger C, Bäuerlein C, Höbartner C (2020) Supramolecular fluorescence resonance energy transfer in nucleobase-modified fluorogenic RNA aptamers. Angew Chem Int Ed 59:6760–6764
Rieder R, Höbartner C, Micura R (2009) Enzymatic ligation strategies for the preparation of purine riboswitches with site-specific chemical modifications. Methods Mol Biol 540:15–24
Horn T, Urdea MS (1986) A chemical 5′-phosphorylation of oligodeoxyribonucleotides. DNA 5:421–426
Sillen A, Engelborghs Y (1998) The correct use of “average” fluorescence parameters. Photochem Photobiol 67:475–486
Lakowicz JR (2010) Principles of fluorescence spectroscopy. Springer, New York, NY
Füchtbauer AF, Wranne MS, Bood M et al (2019) Interbase FRET in RNA: from A to Z. Nucleic Acids Res 47:9990–9997
Cantor CR, Saenger W (1984) Principles of nucleic acid structure. Springer, New York, NY
England TE, Uhlenbeck OC (1978) Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry 17:2069–2076
Romaniuk E, Mclaughlin LW, Neilson T et al (1982) The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur J Biochem 125:639–643
Acknowledgments
This work was supported by the European Research Council (ERC-CoG grant No. 682586 to C.H.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Steinmetzger, C., Höbartner, C. (2023). Probing of Fluorogenic RNA Aptamers via Supramolecular Förster Resonance Energy Transfer with a Universal Fluorescent Nucleobase Analog. In: Mayer, G., Menger, M.M. (eds) Nucleic Acid Aptamers. Methods in Molecular Biology, vol 2570. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2695-5_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2695-5_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2694-8
Online ISBN: 978-1-0716-2695-5
eBook Packages: Springer Protocols