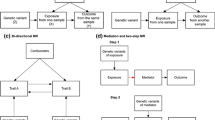
Abstract
Making drug development more efficient by identifying promising drug targets can contribute to resource savings. Identifying promising drug targets using human genetic approaches can remove barriers related to translation. In addition, genetic information can be used to identify potentially causal relationships between a drug target and disease. Mendelian randomization (MR) is a class of approaches used to identify causal associations between pairs of genetically predicted traits using data from human genetic studies. MR can be used to prioritize candidate drug targets by predicting disease outcomes and adverse events that could result from the manipulation of a drug target. The theory behind MR is reviewed, including a discussion of MR assumptions, different MR analytical methods, tests for violations of assumptions, and MR methods that can be robust to some violations of MR assumptions. A protocol to perform two-sample MR (2SMR) with summary genome-wide association study (GWAS) results is described. An example of 2SMR examining the causal relationship between low-density lipoprotein (LDL) and coronary artery disease (CAD) is provided as an illustration of the protocol.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
DiMasi JA, Feldman L, Seckler A, Wilson A (2010) Trends in risks associated with new drug development: success rates for investigational drugs. Clin Pharmacol Ther 87:272–277
Hay M, Thomas DW, Craighead JL et al (2014) Clinical development success rates for investigational drugs. Nat Biotechnol 32:40–51
Plenge RM, Scolnick EM, Altshuler D (2013) Validating therapeutic targets through human genetics. Nat Rev Drug Discov 12:581–594
Ioannidis JPA (2012) Extrapolating from animals to humans. Sci Transl Med 4:151ps15
Perel P, Roberts I, Sena E et al (2007) Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ 334:197
Smith GD, Ebrahim S (2002) Data dredging, bias, or confounding. BMJ 325:1437–1438
Lash TL, VanderWeele TJ, Haneause S, Rothman K (2020) Modern epidemiology, 4th edn. Wolters Kluwer Health
Forshed J (2017) Experimental design in clinical ‘omics biomarker discovery. J Proteome Res 16:3954–3960
Epidemiology for the uninitiated. https://www.bmj.com/about-bmj/resources-readers/publications/epidemiology-uninitiated. Accessed 10 Oct 2021
Bennett DA, Holmes MV (2017) Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart 103:1400–1407
Savitz DA (2014) Invited commentary: interpreting associations between exposure biomarkers and pregnancy outcome. Am J Epidemiol 179:545–547
Schadt EE, Lamb J, Yang X et al (2005) An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37:710–717
Smith GD, Ebrahim S (2003) “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32:1–22
Fang H, ULTRA-DD Consortium, De Wolf H, et al (2019) A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet 51:1082–1091
Plenge RM (2019) Priority index for human genetics and drug discovery. Nat Genet 51:1073–1075
Estrada K, Froelich S, Wuster A et al (2021) Identifying therapeutic drug targets using bidirectional effect genes. Nat Commun 12:2224
Sanseau P, Agarwal P, Barnes MR et al (2012) Use of genome-wide association studies for drug repositioning. Nat Biotechnol 30:317–320
Nelson MR, Tipney H, Painter JL et al (2015) The support of human genetic evidence for approved drug indications. Nat Genet 47:856–860
Smith GD, Ebrahim S (2004) Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33:30–42
Maciejewski ML, Brookhart MA (2019) Using instrumental variables to address bias from unobserved confounders. JAMA 321:2124–2125
Angrist JD, Krueger AB (1992) The effect of age at school entry on educational attainment: an application of instrumental variables with moments from two samples. J Am Stat Assoc 87:328–336
Abifadel M, Varret M, Rabès J-P et al (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34:154–156
Cohen J, Pertsemlidis A, Kotowski IK et al (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37:161–165
Kotowski IK, Pertsemlidis A, Luke A et al (2006) A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet 78:410–422
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354:1264–1272
Davey Smith G, Hemani G (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23:R89–R98
Haycock PC, Burgess S, Wade KH et al (2016) Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr 103:965–978
Zheng J, Baird D, Borges M-C et al (2017) Recent developments in mendelian randomization studies. Curr Epidemiol Rep 4:330–345
Pierce BL, Burgess S (2013) Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 178:1177–1184
Tchetgen Tchetgen EJ, Walter S, Glymour MM (2013) Commentary: building an evidence base for Mendelian randomization studies: assessing the validity and strength of proposed genetic instrumental variables. Int J Epidemiol 42:328–331
Brion MJ, Shakhbazov K, Visscher PM (2013) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42:1497–1501
Buniello A, MacArthur JAL, Cerezo M et al (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47:D1005–D1012
GTEx Consortium (2020) The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369:1318–1330
Hemani G, Zheng J, Elsworth B et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife 7. https://doi.org/10.7554/eLife.34408
Taylor K, Davey Smith G, Relton CL et al (2019) Prioritizing putative influential genes in cardiovascular disease susceptibility by applying tissue-specific Mendelian randomization. Genome Med 11:6
Deelen J, Evans DS, Arking DE et al (2019) A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun 10:3669
Baird DA, Liu JZ, Zheng J et al (2021) Identifying drug targets for neurological and psychiatric disease via genetics and the brain transcriptome. PLoS Genet 17:e1009224
Millard LAC, Davies NM, Timpson NJ et al (2015) MR-PheWAS: hypothesis prioritization among potential causal effects of body mass index on many outcomes, using Mendelian randomization. Sci Rep 5:16645
Evans DM, Brion MJ, Paternoster L et al (2013) Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet 9:e1003919
Zheng J, Haberland V, Baird D et al (2020) Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet 52:1122–1131
Richardson TG, Hemani G, Gaunt TR et al (2020) A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun 11:185
Walker VM, Davey Smith G, Davies NM, Martin RM (2017) Mendelian randomization: a novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol 46:2078–2089
Gill D, Georgakis MK, Walker VM et al (2021) Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res 6:16
Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet 379:1214–1224
Ferreira RC, Freitag DF, Cutler AJ et al (2013) Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet 9:e1003444
Mokry LE, Zhou S, Guo C et al (2019) Interleukin-18 as a drug repositioning opportunity for inflammatory bowel disease: a Mendelian randomization study. Sci Rep 9:9386
Schmidt AF, Holmes MV, Preiss D et al (2019) Phenome-wide association analysis of LDL-cholesterol lowering genetic variants in PCSK9. BMC Cardiovasc Disord 19:240
Thomas DC, Lawlor DA, Thompson JR (2007) Re: estimation of bias in nongenetic observational studies using “Mendelian triangulation” by Bautista et al. Ann Epidemiol 17:511–513
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665
Global Lipids Genetics Consortium, Willer CJ, Schmidt EM et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45:1274–1283
Nikpay M, Goel A, Won H-H et al (2015) A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47:1121–1130
Chang CC, Chow CC, Tellier LC et al (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7
(2005) SNP FAQ archive. The dbSNP mapping process. In: National Center for Biotechnology Information (US), 2005–. https://www.ncbi.nlm.nih.gov/books/NBK573560/
McLaren W, Gil L, Hunt SE et al (2016) The ensembl variant effect predictor. Genome Biol 17:122
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525
Hartwig FP, Davey Smith G, Bowden J (2017) Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46:1985–1998
Bowden J, Davey Smith G, Haycock PC, Burgess S (2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40:304–314
Bowden J, Del Greco MF, Minelli C et al (2017) A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 36:1783–1802
Acknowledgments
This work was supported by NIH U24AG051129.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Evans, D.S. (2022). Target Discovery for Drug Development Using Mendelian Randomization. In: Yan, Q. (eds) Pharmacogenomics in Drug Discovery and Development. Methods in Molecular Biology, vol 2547. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2573-6_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2573-6_1
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2572-9
Online ISBN: 978-1-0716-2573-6
eBook Packages: Springer Protocols