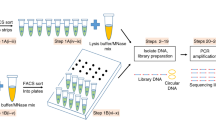
Abstract
Distinct nuclear structures and bodies are involved in genome intranuclear positioning. Measuring proximity and relative distances of genomic loci to these nuclear compartments, and correlating this chromosome intranuclear positioning with epigenetic marks and functional readouts genome-wide, will be required to appreciate the true extent to which this nuclear compartmentalization contributes to regulation of genome functions. Here we present detailed protocols for TSA-seq, the first sequencing-based method for estimation of cytological proximity of chromosomal loci to spatially discrete nuclear structures, such as nuclear bodies or the nuclear lamina. TSA-seq uses Tyramide Signal Amplification (TSA) of immunostained cells to create a concentration gradient of tyramide–biotin free radicals which decays exponentially as a function of distance from a point-source target. Reaction of these free radicals with DNA deposits tyramide–biotin onto DNA as a function of distance from the point source. The relative enrichment of this tyramide-labeled DNA versus input DNA, revealed by DNA sequencing, can then be used as a “cytological ruler” to infer relative, or even absolute, mean chromosomal distances from immunostained nuclear compartments. TSA-seq mapping is highly reproducible and largely independent of the target protein or antibody choice for labeling a particular nuclear compartment. Our protocols include variations in TSA labeling conditions to provide varying spatial resolution as well as enhanced sensitivity. Our most streamlined protocol produces TSA-seq spatial mapping over a distance range of ~1 micron from major nuclear compartments using ~10–20 million cells.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Takizawa T, Meaburn KJ, Misteli T (2008) The meaning of gene positioning. Cell 135(1):9–13. https://doi.org/10.1016/j.cell.2008.09.026
Bickmore WA (2013) The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14:67–84. https://doi.org/10.1146/annurev-genom-091212-153515
Chen Y, Zhang Y, Wang Y et al (2018) Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217:4025–4048. https://doi.org/10.1083/jcb.201807108
Zhang L, Zhang Y, Chen Y et al (2020) TSA-seq reveals a largely conserved genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes. Genome Res 31:251–264. https://doi.org/10.1101/gr.266239.120
Guelen L, Pagie L, Brasset E et al (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453(7197):948–951. https://doi.org/10.1038/nature06947
Kind J, Pagie L, de Vries SS et al (2015) Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163:134–147. https://doi.org/10.1016/j.cell.2015.08.040
van Steensel B, Belmont AS (2017) Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169(5):780–791. https://doi.org/10.1016/j.cell.2017.04.022
Hall LL, Smith KP, Byron M et al (2006) Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288(7):664–675. https://doi.org/10.1002/ar.a.20336
Chen Y, Belmont AS (2019) Genome organization around nuclear speckles. Curr Opin Genet Dev 55:91–99. https://doi.org/10.1016/j.gde.2019.06.008
Spector DL (2001) Nuclear domains. J Cell Sci 114(16):2891
Spector DL (2006) SnapShot: cellular bodies. Cell 127(5):1071. https://doi.org/10.1016/j.cell.2006.11.026
Ferrai C, de Castro IJ, Lavitas L et al (2010) Gene positioning. Cold Spring Harb Perspect Biol 2(6):a000588. https://doi.org/10.1101/cshperspect.a000588
Geyer PK, Vitalini MW, Wallrath LL (2011) Nuclear organization: taking a position on gene expression. Curr Opin Cell Biol 23(3):354–359. https://doi.org/10.1016/j.ceb.2011.03.002
Feuerborn A, Cook PR (2015) Why the activity of a gene depends on its neighbors. Trends Genet 31(9):483–490. https://doi.org/10.1016/j.tig.2015.07.001
Feric M, Vaidya N, Harmon Tyler S et al (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. https://doi.org/10.1016/j.cell.2016.04.047
Yamazaki T, Souquere S, Chujo T et al (2018) Functional domains of NEAT1 architectural lncRNA induce Paraspeckle assembly through phase separation. Mol Cell 70(6):1038–1053e1037. https://doi.org/10.1016/j.molcel.2018.05.019
Rai AK, Chen JX, Selbach M et al (2018) Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559(7713):211–216. https://doi.org/10.1038/s41586-018-0279-8
Strom AR, Brangwynne CP (2019) The liquid nucleome - phase transitions in the nucleus at a glance. J Cell Sci 132(22):jcs235093. https://doi.org/10.1242/jcs.235093
Hondele M, Sachdev R, Heinrich S et al (2019) DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573(7772):144–148. https://doi.org/10.1038/s41586-019-1502-y
Sabari BR, Dall'Agnese A, Boija A et al (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361(6400):eaar3958. https://doi.org/10.1126/science.aar3958
Cho W-K, Spille J-H, Hecht M et al (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361(6400):412. https://doi.org/10.1126/science.aar4199
Boija A, Klein IA, Sabari BR et al (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175(7):1842–1855e1816. https://doi.org/10.1016/j.cell.2018.10.042
Boehning M, Dugast-Darzacq C, Rankovic M et al (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 25(9):833–840. https://doi.org/10.1038/s41594-018-0112-y
Lu H, Yu D, Hansen AS et al (2018) Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558(7709):318–323. https://doi.org/10.1038/s41586-018-0174-3
Guo YE, Manteiga JC, Henninger JE et al (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572(7770):543–548. https://doi.org/10.1038/s41586-019-1464-0
Saitoh N, Spahr CS, Patterson SD et al (2004) Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 15(8):3876–3890. https://doi.org/10.1091/mbc.E04-03-0253
Galganski L, Urbanek MO, Krzyzosiak WJ (2017) Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45(18):10350–10368. https://doi.org/10.1093/nar/gkx759
Dopie J, Sweredoski MJ, Moradian A et al (2020) Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins. J Cell Biol 219(9):e201910207. https://doi.org/10.1083/jcb.201910207
Su J-H, Zheng P, Kinrot SS et al (2020) Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182(6):1641–1659.e1626. https://doi.org/10.1016/j.cell.2020.07.032
Takei Y, Yun J, Ollikainen N et al (2020) Global architecture of the nucleus in single cells by DNA seqFISH+ and multiplexed immunofluorescence. bioRxiv:2020.2011.2029.403055. https://doi.org/10.1101/2020.11.29.403055
Robinett CC, Straight A, Li G et al (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135(6):1685–1700. https://doi.org/10.1083/jcb.135.6.1685
Hepperger C, Otten S, von Hase J et al (2007) Preservation of large-scale chromatin structure in FISH experiments. Chromosoma 116(2):117–133. https://doi.org/10.1007/s00412-006-0084-2
Landt SG, Marinov GK, Kundaje A et al (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22(9):1813–1831. https://doi.org/10.1101/gr.136184.111
Vogel MJ, Peric-Hupkes D, van Steensel B (2007) Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2(6):1467–1478. https://doi.org/10.1038/nprot.2007.148
Skene PJ, Henikoff S (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6:e21856. https://doi.org/10.7554/eLife.21856
Skene PJ, Henikoff JG, Henikoff S (2018) Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13(5):1006–1019. https://doi.org/10.1038/nprot.2018.015
Kaya-Okur HS, Wu SJ, Codomo CA et al (2019) CUT&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10(1):1930. https://doi.org/10.1038/s41467-019-09982-5
van Schaik T, Vos M, Peric-Hupkes D et al (2020) Cell cycle dynamics of lamina-associated DNA. EMBO Rep 21(11):e50636. https://doi.org/10.15252/embr.202050636
Pickersgill H, Kalverda B, de Wit E et al (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38(9):1005–1014. https://doi.org/10.1038/ng1852
Briand N, Collas P (2020) Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21(1):85. https://doi.org/10.1186/s13059-020-02003-5
Ilik İA, Malszycki M, Lübke AK et al (2020) SON and SRRM2 are essential for nuclear speckle formation. Elife 9:e60579. https://doi.org/10.7554/eLife.60579
Fei J, Jadaliha M, Harmon TS et al (2017) Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 130(24):4180. https://doi.org/10.1242/jcs.206854
Quinodoz SA, Ollikainen N, Tabak B et al (2018) Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174(3):744–757e724. https://doi.org/10.1016/j.cell.2018.05.024
Quinodoz SA, Bhat P, Ollikainen N et al (2020) RNA promotes the formation of spatial compartments in the nucleus. bioRxiv:2020.2008.2025.267435. https://doi.org/10.1101/2020.08.25.267435
Chen W, Yan Z, Li S et al (2018) RNAs as proximity-labeling media for identifying nuclear speckle positions relative to the genome. iScience 4:204–215. https://doi.org/10.1016/j.isci.2018.06.005
Bobrow MN, Harris TD, Shaughnessy KJ et al (1989) Catalyzed reporter deposition, a novel method of signal amplification: application to immunoassays. J Immunol Methods 125(1–2):279–285
Raap AK, van de Corput MPC, Vervenne RAM et al (1995) Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum Mol Genet 4(4):529–534. https://doi.org/10.1093/hmg/4.4.529
Gao XD, Tu LC, Mir A et al (2018) C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat Methods 15(6):433–436. https://doi.org/10.1038/s41592-018-0006-2
Myers SA, Wright J, Peckner R et al (2018) Discovery of proteins associated with a predefined genomic locus via dCas9–APEX-mediated proximity labeling. Nat Methods 15(6):437–439. https://doi.org/10.1038/s41592-018-0007-1
Fazal FM, Han S, Parker KR et al (2019) Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178(2):473–490e426. https://doi.org/10.1016/j.cell.2019.05.027
Kurihara M, Kato K, Sanbo C et al (2020) Genomic profiling by ALaP-Seq reveals transcriptional regulation by PML bodies through DNMT3A exclusion. Mol Cell 78(3):493–505.e498. https://doi.org/10.1016/j.molcel.2020.04.004
Tran JR, Paulson DI, Moresco JJ et al (2021) An APEX2 proximity ligation method for mapping interactions with the nuclear lamina. J Cell Biol 220(1):e202002129. https://doi.org/10.1083/jcb.202002129
Hopman AHN, Ramaekers FCS, Speel EJM (1998) Rapid synthesis of biotin-, Digoxigenin-, Trinitrophenyl-, and Fluorochrome-labeled Tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem 46(6):771–777. https://doi.org/10.1177/002215549804600611
http://wiki.xenbase.org/xenwiki/index.php/Flourescin_Tyramide_Synthesis
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Acknowledgments
We thank Drs. William Brieher, Brian Freeman, K.V. Prasanth, and Lisa Stubbs (UIUC, Urbana, IL) for helpful suggestions in developing these protocols. We also thank Belmont laboratory members for reagents and suggestions. We thank the UIUC Biotechnology center for guidance with DNA sonication and sequencing library preparation. We thank Drs. Jian Ma, Bas van Steensel, David Gilbert, and Huimin Zhao from the Belmont 4DN NOFIC U54 Center and other members of the 4D-Nucleome Consortium for helpful suggestions and feedback. This work was supported by National Institutes of Health grants R01GM58460 (ASB) and U54 DK107965 (ASB).
Author contributions: ASB conceptualized the TSA-seq idea and supervised the development of TSA-seq 1.0 and 2.0. YC developed TSA-seq 1.0 with protocols for tyramide–biotin labeling, TSA cell labeling, genomic DNA purification and fragmentation, dot blot and biotinylated DNA bead pulldown, contributed by LZ for Condition 3 (DTT). LZ developed TSA-seq 2.0, added protocols for TSA adherent cell labeling and Drosophila DNA spike in controls, and optimized protocols/methods for TSA labeling, genomic DNA purification and fragmentation, dot blot, and sequencing library construction. LZ, ASB, and YC prepared the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Zhang, L., Chen, Y., Belmont, A.S. (2022). Measuring Cytological Proximity of Chromosomal Loci to Defined Nuclear Compartments with TSA-seq. In: Sexton, T. (eds) Spatial Genome Organization. Methods in Molecular Biology, vol 2532. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2497-5_8
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2497-5_8
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2496-8
Online ISBN: 978-1-0716-2497-5
eBook Packages: Springer Protocols