Abstract
Recently, it has been proposed to switch molecular markers to near-infrared (NIR) spectra for inferring relationships between individuals and further performing phenomic selection (PS), analogous to genomic selection (GS). The PS concept is similar to genomic-like omics-based (GLOB) selection, in which molecular markers are replaced by endophenotypes, such as metabolites or transcript levels, except that the phenomic information obtained for instance by near-infrared spectroscopy (NIRS ) has usually a much lower cost than other omics. Though NIRS has been routinely used in breeding for several decades, especially to deal with end-product quality traits, its use to predict other traits of interest and further make selections is new. Since the seminal paper on PS , several publications have advocated the use of spectral acquisition (including NIRS and hyperspectral imaging) in plant breeding towards PS , potentially providing a scope of what is possible. In the present chapter, we first come back to the concept of PS as originally proposed and provide a classification of selected papers related to the use of phenomics in breeding. We further provide a review of the selected literature concerning the type of technology used, the preprocessing of the spectra, and the statistical modeling to make predictions. We discuss the factors that likely affect the efficiency of PS and compare it to GS in terms of predictive ability. Finally, we propose several prospects for future work and application of PS in the context of plant breeding.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Plant breeding
- Phenomic selection (PS)
- Genomic-like omics-based (GLOB) selection
- Genomic selection (GS)
- Near-infrared spectroscopy (NIRS )
- Hyperspectral imaging
1 Introduction: The Concept of Phenomic Selection and its Relationship with Other Uses of Spectra in Breeding
In their recent publication , Rincent et al. [1] proposed to replace genomic information by phenomic information, such as near-infrared (NIR) spectra, to predict quantitative traits and further perform what they coined “phenomic selection” (PS ). The use of spectroscopy in agriculture and plant breeding is not novel; however, its use as an alternative to molecular markers to build relationship matrices and further predict individual performances in the context of selection is new. PS is similar to genomic-like omics-based (GLOB) selection, in which molecular markers are replaced by endophenotypes such as transcriptomics, metabolomics, or any other omics [2,3,4,5,6,7,8]. Endophenotypes are generally measured once and for all in controlled conditions, and used to build a predictive model for field agronomic traits. This approach is thus similar to genomic selection (GS) with genotyping in the lab replaced by endophenotypic characterization in controlled conditions. In these publications, models such as G-BLUP or multi-BLUP were generally used with the kinship matrix replaced by relationship matrices estimated with the omics data, and the predictive abilities obtained were generally similar and sometimes higher than those obtained with GS . Even if endophenotypic characterization remains costly, improved efficiency to capture non-additive effects (epistasis, genotype by environment interaction, GEI) can be highly valuable. To decrease and scale both cost and throughput, Rincent et al. [1] proposed to replace genotyping or omics characterization by NIRS .
They illustrated that NIR spectra were indeed able to capture genetic similarities, and thus resulted in accurate predictions, even for traits unrelated to the tissue on which NIR spectra were measured. A GLOB selection approach based on NIR spectra acquired in one given environment was for instance able to accurately predict yield in other environments, as long as the calibration set was phenotyped in these environments. Note that PS , and in particular GLOB selection, is radically different from the classical NIRS use. In the classical use, NIRS predicts the chemical composition of the analyzed tissue. In PS and GLOB selection, NIR spectra (or other phenomics data) are used to capture the genetic similarities between the genotypes, which allows accurate predictions of any polygenic trait.
Spectroscopy techniques, such as NIRS , measure the emission or reflection of light on a sample for a given wavenumber range, e.g., for NIR from 780 nm to 2500 nm. Various chemical bonds absorb light at different wavelengths, and this can be used in a quantitative manner. NIRS provides a non-destructive and high-throughput measurement of living samples (where water absorbance bands do not overlap) as well as dried or crushed tissues. Absorption or reflectance values at a given wavelength are proportional to molecule concentrations, as depicted in the Beer–Lambert law. Thus, spectra variations are due to the combination of molecules in the tissue and their respective absorption bands. This chemical property has been widely used in agriculture and forestry in many species to predict traits of interest, such as those related to grain composition for cereals [9], milk composition for dairy cattle [10], wood properties for forest trees [11], quality traits for fruits and vegetables [12].
Apart from this classical use of NIRS , which can be exploited in breeding to make selections on traits related to end-product composition, other uses of NIRS in breeding have been reported in the literature, which can be classified into four main categories as proposed in Table 1: NIR spectra as proxy of the target trait, NIR spectra as a secondary trait, PS within environments, and PS across environments as a particular case of GLOB selection. In the following paragraphs, we provide a definition of each of these four categories.
The first category concerns the use of NIR spectra as a proxy of complex traits such as grain yield, water stress, or chlorophyll content with vegetation indices (VIs), which are based on a few wavelength bands. Several indices were successively developed across years, the most famous one being NDVI (normalized difference vegetation index). These indices are strongly correlated with photosynthetic activity and sometimes with yield [13] and are still widely used to reflect biomass or yield of the analyzed plants or plots [14,15,16,17,18]. The advantage here of using NIR spectra as a proxy trait is to avoid expensive phenotyping and to enable indirect selection of target traits before harvest.
In the second category, we have gathered studies that combined NIRS measurements with molecular markers to increase the accuracy of genomic prediction. In that case, NIR spectrum was considered as a secondary trait to be associated with the target trait in a multivariate prediction model [19,20,21]. It is worth mentioning that such studies report two distinct examples of application: one based on canopy reflectance [19, 21], which relates to the overall plant health; and the other based on grain NIRS [20], which relates to the end-product or cumulative energy accumulation of the plant. In any case, this approach is particularly valuable when the predicted set is phenotyped with NIRS (trait-assisted prediction). Other authors have used NIR measurements to specifically account for GEI in the genomic prediction model [22, 23], but this requires NIRS data for each environment to be measured.
The third category includes studies that have investigated using NIR spectra to build a (hyper)spectral relationship matrix between plots/individuals, referred to as H matrix, and integrated it in the prediction equation with or without the genomic relationship matrix (kinship), referred to as G matrix [22,23,24,25]. The NIRS-based similarity between two plots/individuals H(i, j) can be estimated with the following formula:
with S(i, k) the preprocessed absorbance or reflectance (see hereafter) measured on the ith plot/individual for the kth wavelength. S is centered and scaled for each wavelength and has the dimension n (number of plots/individuals) times nw (number of wavelengths). In matrix notation \( H=\frac{SS^{\prime }}{n_w}. \) This PS approach resulted in promising accuracies; however, in these publications, predictions were still made for a specific site-year trial, and so NIR spectra have to be measured on each plot in each environment, with the strong limitation that all varieties have to be grown in each environment.
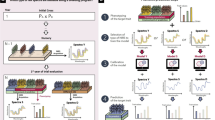
Thus, we propose here a fourth category to further make a distinction between PS applied to the plots on which NIR spectra were acquired (previous category) and PS predictions across environments with NIR spectra measured in a reference site only, a particular case of what we called GLOB selection (scenario S2 in Rincent et al. [1] and Fig. 1). In GLOB selection, we suppose that NIRS captures genetic similarities, which means that spectra acquired in any environment can be useful to make prediction in a particular environment. These external spectra can be (but not necessarily) combined with the spectra potentially collected in the predicted environment. The derived H similarity matrix is then used in replacement of the G matrix in the classical GS models. To date, we have found only two publications (Rincent et al. [1], Lane et al. [26]), that performed GLOB selection with NIRS , and they both resulted in accurate predictions, often more accurate than GS and with dramatically reduced costs. These publications and the results obtained with GLOB selection using other kinds of omics prove that variations other than DNA markers can efficiently capture genetic similarities between genotypes and result in accurate predictions.
Prediction of a target trait for the selection candidates in different environments with GLOB selection. Training population is phenotyped for the target traits (e.g., productivity) in the target environments. Omics or phenomic data (e.g., NIR spectra) are collected on both the candidate and the training individuals in a same reference environment, for capturing a genetic similarity between individuals. Genotypic values of the selection candidates are predicted for the target traits in each target environment
2 Literature Review on the Use of Spectra in Selection
Following previous definitions, we have selected a number of papers illustrating each of the four categories of use of NIRS in the context of breeding. The main features of these papers are summarized in Table 1, and further presented and detailed hereafter, with respect to the type of technology employed to obtain spectra, the statistical pretreatments of the spectra, and the statistical model applied for phenotype prediction. We also provide in Table 2 a comparison of the relative performances of phenomic and genomic predictions for the very few papers which enable such a comparison. Finally, we discuss the factors that affect the predictive ability of PS .
2.1 Types of Technology
Traditionally, NIRS measurements are conducted in laboratories under controlled conditions for either dried vegetative tissue (e.g., forages) or dried reproductive tissue (e.g., grain). This kind of data displays many advantages: measurements are robust, low cost and routinely applied by breeders to predict quality traits. There are also a number of disadvantages, there is substantial extra effort needed to bring these materials from the field to the lab, and to dry them so that water absorbance (which overlaps other chemical bond absorbance) is minimized. In these laboratory conditions, spectra are constituted of many wavelengths possibly from the visible and near infrared (400 to 2500 nm approximately), constituting a dataset of hundreds of variables [9].
With the rise of high-throughput phenotyping, spectrum measurements have benefited from technological developments which enables the direct collection of spectra in the field possibly at several time points, like hyperspectral imaging from Unoccupied Aerial Vehicle (UAV) or direct measurements of fresh material with portable (micro-)spectrometers. Hyperspectral imaging takes images with several wavelengths for each pixel, possibly at multiple time points in the visible and in a small portion of the NIR spectrum. A reflectance measure is attributed to some groups of wavelengths (bands) or to individual wavelengths directly. The measurements at the pixel level can be integrated at the microplot level to characterize a unique variety [17, 22, 24, 25]. Portable (micro-)spectrometers have also been developed to measure the reflectance directly in fields on undestroyed fresh material covering the visible and NIR spectrum [18]. Wavelengths can be used directly as variables in predictive models or they can be derived in several indexes, like VIs [17, 22]. VIs describe vegetation properties by summarizing the information of large amounts of data to facilitate processing of camera and satellite images. However, in Aguate et al. [17] the use of all the hyperspectral bands achieved better prediction than using VI individually.
Technologies used to collect NIR spectrum are numerous, each with advantages and disadvantages. On the one hand, the use of NIR spectrometers in laboratory conditions is a robust method but can be time consuming due to collection and possibly preparation of samples. On the other hand, UAV and portable (micro-)spectrometers are quick techniques to collect NIRS but the number of wavelengths available is usually reduced and measurements can be affected by environmental noise which is harder to control in the field than in the laboratory. Depending on the application, trade-offs must be found between labor intensity, costs, and spectrum quality.
To date, very few studies have tried to compare the predictive ability of different spectrum measurements, especially in the context of plant breeding. Recently, Zgouz et al. [27] have reported a dataset of spectra collected on 60 sugarcane samples with 8 visible/NIR spectrometers including handheld micro-spectrometers. Such a dataset is very useful to compare different tools, although results might be context dependent, i.e., the most accurate model for different traits and species might be obtained with different spectrometers. Still, quantifying the gain or the loss of predictive ability for each technique will be helpful to guide in using one technique rather than another for a specific objective. Other techniques could be used to facilitate measurement, for example to combine hyperspectral images and laboratory spectra. Instead of using a spectrometer to measure samples one by one, hyperspectral images can measure several samples at the same time. This would enhance robustness of spectra collection and reduce time of measurements. Beyond technical issues, it is also important to consider practical organizational questions, such as the period at which spectra are measured, to make sure that the predictions are available before the sowing of the next season.
2.2 Preprocessing NIR Spectra
In ideal conditions, NIR spectra are based on the Beer–Lambert law and the sample absorbance is directly linked to the concentration of chemical compounds of the sample. However, in practice, many factors (independent from the sample composition) will influence the measured absorbance. This is the case for instance of temperature or granulometry, which will deform the final spectrum, biasing spectra comparison. To deal with external effects, a mathematical correction or preprocessing can be applied as illustrated in Fig. 2 for spectra collected on bread wheat grains. Mainly two external effects usually need to be corrected: additive and multiple effects. In additive effects, noise affects spectra irrespectively of the wavelength and usually yields a baseline shift which can be corrected with a detrend [28] or a derivation (Fig. 2c) typically carried out through a Savitzky–Golay filter which consists in a polynomial smoothing [29]. The baseline shift appears when the absorbance increases with the wavelength due to the increased light intensity. Multiplicative effects typically affect spectra differently depending on the wavelength and are usually linked to an increase of the distance crossed by the photons (due to different granulometry for example). They can be corrected by a normalization (Fig. 2b). This effect is present when for low absorbances at a wavelength, the variability is also low and for high absorbances at a wavelength, the variability is high. Other preprocessing techniques have also been proposed to specifically deal with an external parameter known to bias spectra such as temperature or hygrometry. This is the case for instance of the method called External Parameter Orthogonalization (EPO, [30]).
Visualization of different filters applied on a spectrum dataset. Each color represents a bread wheat variety. Spectra were collected on grains with a lab spectrometer NIRS 6500 FOSS. (a) Raw spectra (no preprocessing); (b) normalization (standard normal variate); (c) first derivative of raw spectra; (d) first derivative on normalized spectra
The preprocessing methods briefly introduced in the previous paragraph have been previously developed in the chemometrics literature. This preprocessing is routinely and widely used when applying NIRS in the classical way, i.e., to predict the composition of end-products. In the context of breeding and PS , further preprocessing taken from the breeding literature can be carried out to improve the ability of the spectra to predict genetic values. Such preprocessing includes building a model on the absorbance or reflectance at each wavelength taking into account the effects of the experimental design (e.g., blocks or spatial effects) together with genetic effects to further extract genotypic values [25, 26]. Genotypic values may be BLUEs or BLUPs depending on whether the genotype effect is considered as fixed or random in the model. This preprocessing typically comes from the fact that PS is carried out at the genotype level rather than at the individual or plot level, and consequently one needs to obtain a unique NIRS matrix at the genotype level for model training and prediction. It is interesting to note that if the entire spectrum is considered rather than absorbance or reflectance at given wavelengths, such corrections are related to the orthogonalization approaches from the chemometrics literature. Indeed, recently Ryckeweart et al. [31] proposed to make use of spectra replicates, typically obtained when characterizing plants under genetic trials, to reduce the repeatability error. They developed a new preprocessing technique based on orthogonalization after an ANOVA–simultaneous component analysis (REP-ASCA).
The filters mentioned previously are not an exhaustive list but have been the most commonly used in NIRS chemometric prediction. Preprocessing can be done in numerous ways, as shown across different studies, suggesting that no one standard preprocessing approach exists. We have noticed that PS predictions were influenced by the preprocessing applied on spectra, consequently we recommend testing different filters on a subset of data to cross-validate filters efficiency, before carrying out deeper analysis.
2.3 Statistical Models for Phenotype Prediction
NIRS reflectances or absorbances are quantitative variables, like bi-allelic markers usually coded numerically with allelic dosages, basically all models developed or used in the frame of genomic selection can also be used for PS , from the “infinitesimal” model to Bayesian models with various prior distributions or machine learning methods.
One such reference model for PS is the H-BLUP, similar to G-BLUP but with a similarity matrix (H) estimated with NIRS [1, 23, 25]. Different kernels can be used within such a framework, including Gaussian kernel or arc-cosine kernel [24]. As with molecular markers, this model can be equivalent to a ridge regression on the wavelengths, provided the H matrix is computed accordingly, as demonstrated hereafter. The predictive ability of the H-BLUP model can be measured with cross-validation, as with G-BLUP or other GS models.
The H-BLUP model is defined as: y = μ + u + e, with\( \mathit{\operatorname{var}}(y)=H{\sigma}_u^2+I{\sigma}_e^2 \), and where y is a vector of phenotypes, H is the NIR spectra-based similarity matrix as defined above, μ is the intercept, u and e are random genetic and residual effects, respectively. The RRN-BLUP model (Ridge Regression NIRS BLUP) is defined as: y = μ + Sv + e, with \( \mathit{\operatorname{var}}(y)={SS}^{\prime }{\sigma}_S^2+I{\sigma}_e^2 \), and where S is the matrix of preprocessed, centered, and scaled NIRS as defined above. The mean of y is equal to μ in both models, thus H-BLUP and RRN-BLUP are equivalent if \( H{\sigma}_u^2=S{S}^{\prime {\sigma}_S^2} \), which is for instance the case when \( H=\frac{SS^{\prime }}{n_w} \) and \( {\sigma}_S^2=\frac{\sigma_u^2}{n_w} \).
Functional regression models seem particularly interesting for PS , as they model the linear trend of the spectra [22]. Different kinds of functional regressions were proposed such as functional B-Spline, functional Fourier [22], and Bayesian functional [32]. H-BLUP and functional regression models have proven to yield accurate predictions while reducing computational time by diminishing the number of parameters to estimate. This could be important if several spectra from different environments are available, resulting in a high number of predictors.
Partial Least Squares (PLS) regression, classically used in chemometrics, or variable selection approaches (such as LASSO or BayesB) can also be used to tackle multicollinearity and high dimensionality. PLS regression consists of condensing the information contained across all wavelengths into a few orthogonal variables that maximize the covariation between the predictor matrix and the response variable. In LASSO and BayesB, it is assumed that only a portion of the variables has an effect on the trait. Variable selection seems promising for PS , because the spectrum could be restricted to its most heritable parts [25, 33]. However, it should be noted that the preselection of wavelengths using vegetation indices or with knowledge on the genomic heritability of the wavelengths generally result in lower prediction accuracies than when using the full spectrum [17, 25].
In GS , the choice of the prediction model can be guided by the expected genetic architecture of the predicted trait. The choice of a PS model adapted to a given trait cannot yet rely on such assumptions, and it is not clear how the optimal prediction model can be related to the trait characteristics. The various models tested in the literature sometimes resulted in contrasted prediction accuracies, but in general, sophisticated models were not better than a simple H-BLUP. Models relying on a mixture of distributions such as BayesR [34] are accurate for contrasted genetic architecture in GS ; it would be interesting to test them in PS . In any case, alternative prediction models should be compared using cross-validations within the calibration set.
Contrary to molecular markers in GS , in PS several spectra corresponding to different replicates of genotypes possibly across different environments can be available to build predictive models. In this case, one possibility for calibration is to test each spectrum in order to determine the one which yields the most accurate predictions. Another possibility is to make use of all the available spectra. Lane et al. [26] proposed in the frame of the H-BLUP model to compute the mean of the relationship matrices calculated from each spectrum individually. It is noteworthy that this proposition is equivalent to computing the relationship matrix from a large combined spectra matrix, providing that the individual spectra matrices have the same number of wavelengths, as shown hereafter.
The similarity matrix HT(i, j) computed with the combined spectra matrix (in which all spectra matrices are included one next to the other) is given by:
ST has dimension n (number of individuals) times nt = nl × nw, with nl the number of spectra (e.g., number of environments in which NIRS was acquired) and nw the number of wavelengths of each spectrum (we consider that all spectra have the same wavelengths). Sud(i, k) is the absorbance or reflectance measured on the ith individual for the kth wavelength in the uth NIR preprocessed spectrum. Sud has the dimension n (number of individuals) times nw (number of wavelengths). Hu(i, j) is the similarity between individuals i and j estimated with one given u spectrum.
2.4 Relative Performance of PS Versus GS
There are very few studies that compare PS (and in particular GLOB selection) with GS (Table 2). Although Lane et al. [26] was one of the two studies that implemented GLOB prediction with spectra following our definition, it could not be included in this comparison because they did not apply GS . Table 2 illustrates that PS and GLOB selection have been mainly implemented on cereal species, probably because of the widespread and routine use of NIR measurements on grains to predict protein content. Krause et al. [23] and Galán et al. [25] reported similar accuracies for GS and PS , while Cuevas et al. [24] showed lower accuracies for PS compared to GS (0.37 and 0.46, respectively). The highest PS accuracy compared with GS was observed in Rincent et al. [1] in wheat. The lowest PS accuracy compared to GS was observed in Rincent et al. [1] in poplar for which NIR spectra were collected on wood for a reduced range of wavelengths. From these data, it is apparent that PS had comparable or higher accuracies than GS in most cases. Even in cases where PS is less accurate than GS , as NIR measurements are high throughput and low cost compared to genotyping, PS could still provide higher genetic gains than GS , as demonstrated in Rincent et al. [1]. In our ongoing research we compared GS and PS at different generations of elite bread wheat selection. We found that PS could be as accurate as GS and even better when applied to early generations. Further work on other species is clearly needed to deepen this comparison and provide valuable information on the factors and conditions (e.g., tissue, environment) that determine the predictive ability of NIRS .
By considerably reducing the costs of implementation, PS is a tool of choice to improve the balance between costs and benefits in comparison with GS . PS would be particularly valuable for orphan crops for which genotyping is expensive, or for major crops for which phenomic data are already routinely collected (e.g., maize and wheat). In the latter case, phenomic prediction already opens new possibilities in existing breeding programs without any additional cost, and with predictive abilities similar to those obtained with genomic prediction [1].
2.5 Factors Affecting PS Predictive Ability
In the past, several kinds of omics were used to make genomic-like predictions with promising results [8]. NIRS captures an integrative signal, and is biologically more difficult to interpret than other omics, which describe each molecule individually (e.g., transcriptomics, proteomics, metabolomics). However, because prediction models do not necessarily need to be interpreted biologically, NIRS can be used to make predictions using “black-box” models.
There are two factors that contribute to the predictive ability and consequently to the success of PS : (1) the ability to capture target trait proxies and (2) the ability to infer genetic relatedness. The former depends on the physiological connectedness between the target trait and the composition and features of the tissue analyzed with NIRS . This is for example the case when NIR spectra collected on wood powder is used to predict wood properties, or when NIR spectra collected on fruits is used to predict fruit composition. In these cases, PS should be nearly equivalent or superior to the traditional way of using NIRS (prediction of the tissue composition), the only difference being that when doing PS we usually work at the genotype level because we aim at ranking and selecting the best genotypes while in the traditional use of NIRS we make predictions at the plot or plant level [26]. However, we could think of more indirect relationships between the target trait and NIR spectra to explain its predictive ability, for instance in wheat the good predictive ability of PS for yield could be due to the fact that NIRS is a very good predictor of grain composition, that is often negatively correlated to yield. This could also be the case for maturity: the spectra are influenced by the maturity of the plants, and this maturity is sometimes correlated to yield [23]. However, it is important to stress that even in the absence of any direct relationship between the predicted trait and the tissue analyzed with NIRS , PS can still be accurate. It was for instance shown in Rincent et al. [1] that NIR spectra collected on leaves in one environment could be used to estimate a covariance matrix resulting in accurate prediction of yield in a completely independent environment. In this particular example, the correlation between yield in the environment in which NIR spectra were obtained, and yield in the predicted environment (by cross-validation within the predicted environment) was as low as 0.16, whereas PS predictive ability was above 0.5. This means that NIRS-derived relationship matrices were able to capture genetic relatedness between lines valuable for predicting yield. This was further demonstrated by the fact that genomic heritability was significant for many wavelengths. A further demonstration could be done with a simulation study, by estimating the predictive ability of PS for traits simulated with genotype data. In this case, the predictive ability of PS averaged over a large number of simulated traits would provide an evaluation of the ability of NIRS to infer genetic relationships for predicting quantitative traits unrelated to the tissue composition.
PS is a recent research topic, and further investigations are required to use it in an optimal way. One can expect that, as for genomic selection, prediction accuracy will be strongly dependent on the target trait and its heritability , as well as the size and composition of the training set. In comparison to GS , prediction accuracy obtained with PS can also be affected by the origin of the spectrum (tissue, environments, kind of sensors). This is similar to the choice of a SNP array and marker filtering in GS , but the effect of the origin of the spectrum appears to be more pronounced. First results suggest that NIR spectra collected under plant stress conditions are more efficient [1, 26], but other experiments are required before it can be understood if this is the rule or the exception. An interesting result is that, in practice, the combination of different NIR spectra (collected on different tissues or different environments) leads to predictive abilities at least as good as those obtained with the best NIR spectrum taken alone ( [1], unpublished results). This means that in some cases, it is not necessary to identify the best conditions to obtain NIR spectra, but simply to aggregate all the spectra collected (e.g., spectra obtained on the same genotypes at the different steps of the breeding program). As shown in the present review, aggregating NIRS matrices prior to computing the H matrix is equivalent to averaging H matrices estimated with individual NIRS matrices and is thus quite straightforward. However, in any case, the choice of the tissue, timing, and sensors could and should also be optimized. We can think that NIR spectra collected on homogeneous, representative samples (leaf powder, seed sample, or flour) are more useful than NIR spectra obtained on a tiny area of raw material. The way NIR spectra are collected should also be optimized in terms of practical feasibility. For instance in wheat, it would be much more feasible to measure NIR spectra during the growing season, than on grain after harvest, because the few weeks between harvest and sowing of the next generation is labor intensive and so NIR spectra acquisition would be difficult during this period.
3 Prospects
As introduced above, there are numerous ways of using PS in breeding. We particularly foresee several applications to be addressed with PS and that we detail hereafter. Some of them are quite direct applications, which can already be deployed in breeding programs (see below Subheadings 3.1–3.6), while others are prospects which deserve research investments prior to their adoption in breeding in a relatively longer term (Subheading 3.7). Although many of the prospects presented here are also shared with GS , we have tried in what follows to underline their specificity with respect to PS .
3.1 Prebreeding: Screening Diversity Collections at Low Cost
Gene banks are a reservoir of genetic diversity in which genes of tolerance to biotic or abiotic stress can be discovered. These collections will become crucial, as the genetic diversity in the breeding program will not be sufficient to face upcoming changes due to the evolution of management practices or climate change. However, the identification of promising genes or individuals require the phenotyping of the collection for the target trait, which could be too expensive given the number of accessions stored in these collections. Yu et al. [35] and Crossa et al. [36] have proposed to run GS to screen these collections. However, this requires genotyping the full collection, which is also expensive considering the large size of the gene banks. Another option would be to measure NIR spectra on each accession, phenotype a subset, and predict the remaining accessions using PS . In most gene banks, accessions are regularly sown in nurseries to produce new seeds, as the germination rate decreases with time. NIRS could thus either be collected on seeds directly in gene banks or on other tissues in these nurseries. The same approach could also be extended to other species like perennials which are usually not conserved as seeds but as living plants in the field.
3.2 Sparse Testing: Experimental Design Optimization in Breeding Programs
For most species, selection candidates are evaluated in multi-environment trials to estimate their stability and productivity in contrasted environments. This is an expensive step as the number of variety/environment combinations can rapidly become very high. To increase the number of environments or varieties with the same costs, one option would be to run a sparse testing design, in which all varieties are evaluated at least in one environment, but with a given proportion of varieties/environments not tested. Sparse testing is sometimes imposed to breeders because part of a trial is accidentally destroyed, or not harvested for some reason. Sparse testing is a scenario for which genomic predictions are particularly accurate, in comparison to predicting a completely new variety or a new environment [37,38,39]. Our proposition is to use PS to predict the unobserved variety/environment combinations. In wheat, this approach is already applicable as NIR spectra are usually collected in nurseries the year before the multi-environment trials. Our first unpublished results on sparse testing show that the prediction accuracy of PS under this scenario can be as high as to the one obtained with GS .
3.3 Combining Reduction of Generation Time (Speed Breeding) and Performance Prediction (PS ) to Increase Genetic Progress
One challenge in breeding is to accelerate programs to quickly release new varieties. Breeding is often constrained to one to two generations per year for annual crops, which limits genetic progress. Several methods have been proposed to reduce generation time, including recently “speed breeding” [40]. Speed breeding consists in controlling photoperiod and temperature to get optimal growing conditions and accelerate the time elapsed from seed to seed, allowing up to six spring wheat generations per year. Thus, a great number of segregation and recombination events can occur in a short time, allowing to rapidly produce varieties combining favorable alleles. PS could have two applications that could work particularly well if combined with speed breeding. The first application could be implemented in the speed breeding process itself. During this process, each plant is unique and is phenotypically quite different from what it would look in the field, which makes direct selection impossible except for phenology, height, and some disease resistance traits. But molecular markers or NIR spectra could be measured on the plants or on the seeds which would allow predicting performance traits using GS [41], or at a lower cost PS . This would considerably increase genetic progress by reducing both the generation interval (speed breeding) and the phenotyping process (partially replaced by predictions). The second application would be to apply PS to reduce the field trials size after the speed breeding process. Thousands of genotypes can be produced by speed breeding, and it would be difficult to phenotype all of them in field experiments (after multiplication in the nurseries). As NIR spectra can be collected at the end of the process or in the nurseries, PS could be used to predict genotype performance in the nurseries or with sparse testing in the following field experiments as described above.
3.4 GEI Prediction
Unlike molecular markers, NIR spectra are directly influenced by the response of the plants to the environmental conditions. This seems likely to result in a lack of stability of the spectra in different environments. It also may mean that the spectra are able to capture the genetic responses to a given environment (GEI), as illustrated in Rincent et al. [1], which opens new perspectives of application for PS . This GEI variance could be exploited to enhance predictive ability of local adaptation. One possibility to improve the GEI prediction models is to use NIRS collected in each environment to estimate environment-specific covariance matrices. Krause et al. [23] compared different models (single and multi-kernel) using molecular marker, pedigree, or NIR spectra, to predict wheat grain yield in multi-environment trials. They found that the best multi-kernel integrating GEI was the one with the hyperspectral matrix. Using NIRS enhanced predictive accuracy of GEI compared to models that use molecular markers or pedigree. Similar results were observed by Montesinos-López et al. [22] and Lane et al. [26] where the interaction between NIR bands and environments was integrated in the models. The introduction of NIR information in the prediction model allowed increasing GEI prediction accuracy, which was not the case with molecular markers.
This multi-kernel method requires collecting the spectra of all genotypes in all the environments of the multi-environment trials, which is not possible with classical sparse testing designs. One possibility would be to grow a nursery in parallel of each trial, which would be much cheaper than a real trial. In this case, the trial will be dedicated to phenotype the training population and the nursery to collect spectra on both training and predicted lines. Krause et al. [23] proposed to reduce the size of the microplots, to measure NIR spectra on all lines in all environments using a UAV. The objective is to find a compromise between the number of candidate varieties predicted with PS and the number of training varieties phenotyped for the target traits.
Another potential application would be to use NIR spectra collected in the different environments to estimate similarities between them, as proposed in Jarquin et al. [37] and Heslot et al. [42] with environmental covariates. In comparison to classical environmental covariates (e.g., temperature, hydric balance, radiation), NIRS has the advantage of capturing signals from the plant that could be influenced by the experienced stresses. GEI models enriched with NIRS would be particularly useful to make predictions in multi-environment trials. This application of PS could potentially allow making predictions in new environments (no other phenotype than NIR spectra are collected in these environments) by estimating the similarity between the new environment and the calibration environments. NIR spectra can also be used as a covariate in the predictive model to characterize the GEI. Lane et al. [26] proposed to take into account the GEI by using wavelengths as covariates in the predictive model through functional regression. They predicted yields of known hybrids in an unknown environment and found that taking into account GEI with covariates worked better than regular H-BLUP. The different models tested in these studies underline that NIRS can be a good predictor of GEI, and in many different ways. These results are very promising to enhance predictive ability in the context of multi-environment trials.
3.5 Making Use of Historical NIRS Data in Prediction
In GS , the enrichment of the training set with historical data from multiple environments can improve predictive ability by increasing the size of the training set and by limiting the effect of atypical years that are difficult to predict with reduced and traditional datasets [43]. The use of historical data might be more complicated with PS , as NIR spectra are likely to be more specific to the environment in which they are measured. To estimate the NIR similarity matrix (H) between historical varieties characterized with NIRS over successive years (the varieties changing from 1 year to another), the effect of environment and GEI should be accounted for and corrected from the spectra. One option would be to use check varieties for which NIR spectra would be collected each year to determine the transformation from the spectra collected in one environment to the spectra collected in another environment.
3.6 The Case of Perennials
Genetic resources for perennials are usually conserved as collections in nurseries. These collections typically include individuals from different species, populations, genotypes as well as clones. In this case, we foresee several potential applications of PS . First, as already mentioned in the case of gene banks, NIRS could be used to rapidly screen these resources for target traits that are typically difficult to evaluate on a large number of individuals, such as fruit quality or phenology. Second, in a more advanced breeding context, one could imagine that progenies from controlled crosses could be planted near a well-characterized reference panel of widely cultivated varieties. NIRS data could be collected at the same time on both reference and candidate individuals to limit environmental heterogeneity between NIRS matrices from training and test sets, and a model could be trained by using phenotypic data potentially obtained in other sites for the reference panel in order to predict the performance of the candidates in these particular sites. This scenario corresponds to GLOB prediction, which proved to be quite efficient for some traits in wheat and poplar in Rincent et al. [1].
3.7 Other Applications
3.7.1 Genotype Inference
Whalen et al. [44] described a new application of NIRS for breeding beyond PS : the use of High-Throughput Phenotypes (HTP) such as NIRS to infer genotypes. They illustrated their approach with simulations, in which spectra resulted from an additive genetic model, with 100 QTLs per chromosome and heritability of wavelength ranging between 0.1 and 0.7. From that, they fit a model, which links the segregation states at each locus with the HTP, for the training population. Then, this model was applied to predict the segregation states of non-genotyped individuals. They concluded that under certain conditions it was possible to infer the genotypes of individuals derived from biparental crosses. The HTP-enabled genomic prediction (with genotypes inferred from HTP data) yielded higher accuracies than PS , the best accuracies being from classic genomic prediction models with real genotypes. However, it is important to note that PS does not only capture additive genetic effects, but also epistatic and GEI. This advantage of PS over GS could not be illustrated here, as only additive effects were simulated. Nevertheless this application seems very promising for breeding, especially for species with high genotyping cost. Furthermore, as PS relies on global relatedness between genotypes derived from NIRS , it could be difficult to predict traits with mono- or oligogenic genetic architecture. Hence, this transition proposed by Whalen et al. [44] from NIRS to marker data allows to apply GS but also QTL detection. This shows that the use of NIRS instead of genotyping in breeding may be adapted to contrasted genetic architectures. A demonstration of its usefulness with real data remains to be conducted.
3.7.2 Hybrid Prediction
As NIR spectrum is a phenotype, it captures both additive and interaction effects. For this reason, it could be possible to use the NIRS covariance matrix to predict hybrid performances, taking into account both general and specific combining abilities. The idea would be to collect NIR spectra on the hybrids to estimate a covariance matrix taking interaction effects into account. One option would be to collect NIRS data on large collections of hybrids in nurseries, and phenotype only part of the hybrids in classical multi-environment trials, possibly with the sparse testing approach described above.
3.7.3 Progeny sorting
In some programs, breeders are interested in quickly characterizing progenies from controlled crosses with respect to their resemblance to their parents. This is typically the case when crossing an established widely used variety with a donor genotype with the aim to introgress particular features from the donor to the variety. An example of this would be the case of grapevine for which a current challenge is to quickly breed varieties with resistance to biotic factors while maintaining some established quality for wine making. In this case when a given variety is crossed with a disease resistant genotype, it is usually quite straightforward using marker-assisted selection to select the progenies that carry disease resistant genes, but for wine quality it is much more complicated. The goal would be to select among the resistant progenies those that are more similar to the parental wine-making variety, which using molecular markers is a very difficult task without considering prior knowledge on the genetic architecture of the traits. In this particular case, NIRS could be useful to provide a distance between the resistant progenies and their parents.
4 Conclusion
We have reviewed the different approaches that have so far been proposed in literature to predict agronomic traits with NIRS , from prediction at the plot/individual level to PS and GLOB selection. NIRS has been intensively used to make predictions at the plot or genotype level, considering NIRS as a secondary trait or as a yield proxy. The originality of PS and GLOB selection as defined in Rincent et al. [1] is that NIR spectra are used in a similar way as molecular markers in GS . They indeed supposed that NIR spectra were able to capture genetic relationships between individuals. PS resulted in good predictive abilities (often similar or even higher than those obtained with GS ), even when the predicted trait was completely independent from the tissue analyzed with NIRS (e.g., different environments). The high throughput and low cost of this approach makes it interesting to increase breeding efficiency in comparison to GS , particularly for species for which genotyping is expensive, or for crops for which NIRS data are already routinely collected in the breeding programs. We have also proposed different promising applications of PS in breeding and prebreeding among which some can readily be applicable, while others require further work in order to test and optimize this approach.
References
Rincent R, Charpentier J-P, Faivre-Rampant P, Paux E, Le Gouis J, Bastien C, Segura V (2018) Phenomic selection is a low-cost and high-throughput method based on indirect predictions: proof of concept on wheat and poplar. G3 (Bethesda) 8:3961–3972. https://doi.org/10.1534/g3.118.200760
Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44:217–220. https://doi.org/10.1038/ng.1033
Feher K, Lisec J, Römisch-Margl L, Selbig J, Gierl A, Piepho H-P, Nikoloski Z, Willmitzer L (2014) Deducing hybrid performance from parental metabolic profiles of young primary roots of maize by using a multivariate Diallel approach. PLoS One 9:e85435. https://doi.org/10.1371/journal.pone.0085435
Ward J, Rakszegi M, Bedő Z, Shewry PR, Mackay I (2015) Differentially penalized regression to predict agronomic traits from metabolites and markers in wheat. BMC Genet 16:19. https://doi.org/10.1186/s12863-015-0169-0
Fernandez O, Urrutia M, Bernillon S, Giauffret C, Tardieu F, Le Gouis J, Langlade N, Charcosset A, Moing A, Gibon Y (2016) Fortune telling: metabolic markers of plant performance. Metabolomics 12:158. https://doi.org/10.1007/s11306-016-1099-1
Xu S, Xu Y, Gong L, Zhang Q (2016) Metabolomic prediction of yield in hybrid rice. Plant J 88:219–227. https://doi.org/10.1111/tpj.13242
Guo Z, Magwire MM, Basten CJ, Xu Z, Wang D (2016) Evaluation of the utility of gene expression and metabolic information for genomic prediction in maize. Theor Appl Genet 129:2413–2427. https://doi.org/10.1007/s00122-016-2780-5
Schrag TA, Westhues M, Schipprack W, Seifert F, Thiemann A, Scholten S, Melchinger AE (2018) Beyond genomic prediction: combining different types of omics data can improve prediction of hybrid performance in maize. Genetics 208:1373–1385. https://doi.org/10.1534/genetics.117.300374
Osborne BG (2006) Applications of near infrared spectroscopy in quality screening of early-generation material in cereal breeding Programmes. J Near Infrared Spectrosc 14:93–101. https://doi.org/10.1255/jnirs.595
Holroyd SE (2013) The use of near infrared spectroscopy on Milk and Milk products. J Near Infrared Spectrosc 21:311–322. https://doi.org/10.1255/jnirs.1055
Tsuchikawa S, Kobori H (2015) A review of recent application of near infrared spectroscopy to wood science and technology. J Wood Sci 61:213–220. https://doi.org/10.1007/s10086-015-1467-x
Nicolaï BM, Beullens K, Bobelyn E, Peirs A, Saeys W, Theron KI, Lammertyn J (2007) Nondestructive measurement of fruit and vegetable quality by means of NIR spectroscopy: a review. Postharvest Biol Technol 46:99–118. https://doi.org/10.1016/j.postharvbio.2007.06.024
Tucker CJ (1979) Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens Environ 8:127–150. https://doi.org/10.1016/0034-4257(79)90013-0
Labus MP, Nielsen GA, Lawrence RL, Engel R, Long DS (2002) Wheat yield estimates using multi-temporal NDVI satellite imagery. Int J Remote Sens 23:4169–4180. https://doi.org/10.1080/01431160110107653
Ferrio JP, Bertran E, Nachit MM, Català J, Araus JL (2004) Estimation of grain yield by near-infrared reflectance spectroscopy in durum wheat. Euphytica 137:373–380. https://doi.org/10.1023/B:EUPH.0000040523.52707.1e
Babar MA, Reynolds MP, van Ginkel M, Klatt AR, Raun WR, Stone ML (2006) Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci 46:1046–1057. https://doi.org/10.2135/cropsci2005.0211
Aguate FM, Trachsel S, Pérez LG, Burgueño J, Crossa J, Balzarini M, Gouache D, Bogard M, de los Campos G (2017) Use of hyperspectral image data outperforms vegetation indices in prediction of maize yield. Crop Sci 57:2517–2524. https://doi.org/10.2135/cropsci2017.01.0007
Hernandez J, Lobos G, Matus I, del Pozo A, Silva P, Galleguillos M (2015) Using ridge regression models to estimate grain yield from field spectral data in bread wheat (Triticum Aestivum L.) grown under three water regimes. Remote Sens 7:2109–2126. https://doi.org/10.3390/rs70202109
Rutkoski J, Poland J, Mondal S, Autrique E, Pérez LG, Crossa J, Reynolds M, Singh R (2016) Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 (Bethesda) 6:2799–2808. https://doi.org/10.1534/g3.116.032888
Hayes BJ, Panozzo J, Walker CK, Choy AL, Kant S, Wong D, Tibbits J, Daetwyler HD, Rochfort S, Hayden MJ, Spangenberg GC (2017) Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor Appl Genet 130:2505–2519. https://doi.org/10.1007/s00122-017-2972-7
Sun J, Rutkoski JE, Poland JA, Crossa J, Jannink J-L, Sorrells ME (2017) Multitrait, random regression, or simple repeatability model in high-throughput phenotyping data improve genomic prediction for wheat grain yield. Plant Genome 10. https://doi.org/10.3835/plantgenome2016.11.0111
Montesinos-López OA, Montesinos-López A, Crossa J, de los Campos G, Alvarado G, Suchismita M, Rutkoski J, González-Pérez L, Burgueño J (2017) Predicting grain yield using canopy hyperspectral reflectance in wheat breeding data. Plant Methods 13:4. https://doi.org/10.1186/s13007-016-0154-2
Krause MR, González-Pérez L, Crossa J, Pérez-Rodríguez P, Montesinos-López O, Singh RP, Dreisigacker S, Poland J, Rutkoski J, Sorrells M, Gore MA, Mondal S (2019) Hyperspectral reflectance-derived relationship matrices for genomic prediction of grain yield in wheat. G3 (Bethesda) 9:1231–1247. https://doi.org/10.1534/g3.118.200856
Cuevas J, Montesinos-López O, Juliana P, Guzmán C, Pérez-Rodríguez P, González-Bucio J, Burgueño J, Montesinos-López A, Crossa J (2019) Deep kernel for genomic and near infrared predictions in multi-environment breeding trials. G3 (Bethesda) 9:2913–2924. https://doi.org/10.1534/g3.119.400493
Galán RJ, Bernal-Vasquez A-M, Jebsen C, Piepho H-P, Thorwarth P, Steffan P, Gordillo A, Miedaner T (2020) Integration of genotypic, hyperspectral, and phenotypic data to improve biomass yield prediction in hybrid rye. Theor Appl Genet 133:3001–3015. https://doi.org/10.1007/s00122-020-03651-8
Lane HM, Murray SC, Montesinos-López OA, Montesinos-López A, Crossa J, Rooney DK, Barrero-Farfan ID, Fuente GNDL, Morgan CLS (2020) Phenomic selection and prediction of maize grain yield from near-infrared reflectance spectroscopy of kernels. Plant Phenome J 3:e20002. https://doi.org/10.1002/ppj2.20002
Zgouz A, Héran D, Barthès B, Bastianelli D, Bonnal L, Baeten V, Lurol S, Bonin M, Roger JM, Bendoula R, Chaix G (2020) Dataset of visible-near infrared handheld and microspectrometers—comparison of the prediction accuracy of sugarcane properties. Data Brief 31:106013. https://doi.org/10.1016/j.dib.2020.106013
Barnes RJ, Dhanoa MS, Lister SJ (1989) Standard Normal variate transformation and De-trending of near-infrared diffuse reflectance spectra. Appl Spectrosc 43:772–777. https://doi.org/10.1366/0003702894202201
Savitzky A, Golay MJ (1964) Smoothing and differentiation of data by simplified least squares procedures. Anal Chem 36:1627–1639. https://doi.org/10.1021/ac60214a047
Roger J-M, Chauchard F, Bellon-Maurel V (2003) EPO–PLS external parameter orthogonalisation of PLS application to temperature-independent measurement of sugar content of intact fruits. Chemom Intell Lab Syst 66:191–204. https://doi.org/10.1016/S0169-7439(03)00051-0
Ryckewaert M, Gorretta N, Henriot F, Marini F, Roger J-M (2020) Reduction of repeatability error for analysis of variance-simultaneous component analysis (REP-ASCA): application to NIR spectroscopy on coffee sample. Anal Chim Acta 1101:23–31. https://doi.org/10.1016/j.aca.2019.12.024
Montesinos-López A, Montesinos-López OA, Cuevas J, Mata-López WA, Burgueño J, Mondal S, Huerta J, Singh R, Autrique E, González-Pérez L, Crossa J (2017) Genomic Bayesian functional regression models with interactions for predicting wheat grain yield using hyper-spectral image data. Plant Methods 13:62. https://doi.org/10.1186/s13007-017-0212-4
Ferragina A, de los Campos G, Vazquez AI, Cecchinato A, Bittante G (2015) Bayesian regression models outperform partial least squares methods for predicting milk components and technological properties using infrared spectral data. J Dairy Sci 98:8133–8151. https://doi.org/10.3168/jds.2014-9143
Erbe M, Hayes BJ, Matukumalli LK, Goswami S, Bowman PJ, Reich CM, Mason BA, Goddard ME (2012) Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J Dairy Sci 95:4114–4129. https://doi.org/10.3168/jds.2011-5019
Yu X, Li X, Guo T, Zhu C, Wu Y, Mitchell SE, Roozeboom KL, Wang D, Wang ML, Pederson GA, Tesso TT, Schnable PS, Bernardo R, Yu J (2016) Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nat Plants 2:16150. https://doi.org/10.1038/nplants.2016.150
Crossa J, Jarquín D, Franco J, Pérez-Rodríguez P, Burgueño J, Saint-Pierre C, Vikram P, Sansaloni C, Petroli C, Akdemir D, Sneller C, Reynolds M, Tattaris M, Payne T, Guzman C, Peña RJ, Wenzl P, Singh S (2016) Genomic prediction of Gene Bank wheat landraces. G3 (Bethesda) 6:1819–1834. https://doi.org/10.1534/g3.116.029637
Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, Piraux F, Guerreiro L, Pérez P, Calus M, Burgueño J, de los Campos G (2014) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet 127:595–607. https://doi.org/10.1007/s00122-013-2243-1
Jarquin D, Howard R, Crossa J, Beyene Y, Gowda M, Martini JWR, Covarrubias Pazaran G, Burgueño J, Pacheco A, Grondona M, Wimmer V, Prasanna BM (2020) Genomic prediction enhanced sparse testing for multi-environment trials. G3 (Bethesda) 10:2725–2739. https://doi.org/10.1534/g3.120.401349
Ly D, Huet S, Gauffreteau A, Rincent R, Touzy G, Mini A, Jannink J-L, Cormier F, Paux E, Lafarge S, Le Gouis J, Charmet G (2018) Whole-genome prediction of reaction norms to environmental stress in bread wheat (Triticum aestivum L.) by genomic random regression. Field Crops Res 216:32–41. https://doi.org/10.1016/j.fcr.2017.08.020
Watson A, Ghosh S, Williams MJ, Cuddy WS, Simmonds J, Rey M-D, Hatta AM, Hinchliffe A, Steed A, Reynolds D, Adamski NM, Breakspear A, Korolev A, Rayner T, Dixon LE, Riaz A, Martin W, Ryan M, Edwards D, Batley J, Raman H, Carter J, Rogers C, Domoney C, Moore G, Harwood W, Nicholson P, Dieters MJ, IH DL, Zhou J, Uauy C, Boden SA, Park RF, BBH W, Hickey LT (2018) Speed breeding is a powerful tool to accelerate crop research and breeding. Nat Plants 4:23–29. https://doi.org/10.1038/s41477-017-0083-8
Watson A, Hickey LT, Christopher J, Rutkoski J, Poland J, Hayes BJ (2019) Multivariate genomic selection and potential of rapid indirect selection with speed breeding in spring wheat. Crop Sci 59:1945–1959. https://doi.org/10.2135/cropsci2018.12.0757
Heslot N, Akdemir D, Sorrells ME, Jannink J-L (2014) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet 127:463–480. https://doi.org/10.1007/s00122-013-2231-5
Sweeney DW, Sun J, Taagen E, Sorrells ME (2019) Genomic selection in wheat. In: Miedaner T, Korzun V (eds) Applications of genetic and genomic research in cereals. Woodhead Publishing, Sawston. https://doi.org/10.1016/B978-0-08-102163-7.00013-2
Whalen A, Gaynor C, Hickey JM (2020) Using high-throughput phenotypes to enable genomic selection by inferring genotypes bioRxiv 2020.02.28.969600. https://doi.org/10.1101/2020.02.28.969600
Acknowledgments
We would like to thank the reviewer and editors for their suggestions and useful remarks on a previous version of this chapter. We also thank Jacques Le Gouis for proposing the “GLOB” denomination.
Funding: The preparation of this chapter was partly supported by: the project OASIs funded by the French Ministry of Agriculture and Food (“fond CASDAR, appel à projets Semences et sélection végétale 2020, projet n° C-2020-5”), the project SelGenVit funded by the French National Research Agency (ANR-19-ECOM-0006), and the BreedWheat project funded by the French National Research Agency under Investment for the Future (ANR-10-BTBR-03).
Partial funding of Charlotte Brault’s PhD was provided by the Association Nationale de la Recherche et de la Technologie (ANRT, grant number 2018/0577), IFV and Inter-Rhône.
Pauline Robert’s PhD was jointly funded by Agri-Obtentions, Florimond-Desprez, and the Association Nationale de la Recherche et de la Technologie (ANRT, grant number 2019/0060).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Robert, P., Brault, C., Rincent, R., Segura, V. (2022). Phenomic Selection: A New and Efficient Alternative to Genomic Selection. In: Ahmadi, N., Bartholomé, J. (eds) Genomic Prediction of Complex Traits. Methods in Molecular Biology, vol 2467. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2205-6_14
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2205-6_14
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2204-9
Online ISBN: 978-1-0716-2205-6
eBook Packages: Springer Protocols