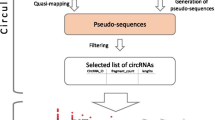
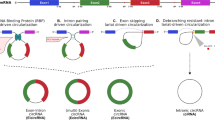
Abstract
Circular RNAs (circRNAs) are a vast class of covalently closed, noncoding RNAs expressed in specific tissues and developmental stages. The molecular, cellular, and pathophysiologic roles of circRNAs are not fully known, but their impact on gene expression programs is beginning to emerge, as circRNAs often associate with RNA-binding proteins and nucleic acids. With rising interest in identifying circRNAs associated with disease processes, it has become particularly important to identify circRNAs in RNA sequencing (RNA-seq) datasets, either generated by the investigator or reported in the literature. Here, we present a methodology to identify and analyze circRNAs in RNA-seq datasets, including those archived in repositories. We elaborate on the unique features of circRNAs that require specialized attention in RNA-seq datasets, the software packages designed for circRNA identification, the ongoing efforts to reconstruct the body of circRNAs starting from unique circularizing junctions, and the interacting factors that can be proposed from putative circRNA body sequences. We discuss the advantages and limitations of the current approaches for high-throughput circRNA analysis from RNA-sequencing datasets and identify areas that would benefit from the development of superior bioinformatic tools.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN (2018) Circular RNA splicing. Adv Exp Med Biol 1087:41–52
Yu CY, Kuo HC (2019) The emerging roles and functions of circular RNAs and their generation. J Biomed Sci 26:29
Kumar L, Shamsuzzama HR, Baghel T, Nazir A (2017) Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol 54:7224–7234
Hu W, Bi ZY, Chen ZL et al (2018) Emerging landscape of circular RNAs in lung cancer. Cancer Lett 427:18–27
Qu S, Yang X, Li X et al (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148
Chen B, Huang S (2018) Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 418:41–50
Xiao MS, Wilusz JE (2019) An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res 47:8755–8769
Panda AC, De S, Grammatikakis I et al (2017) High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 45:e116
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997
Zeng X, Lin W, Guo M, Zou Q (2017) A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13:e1005420
Cheng J, Metge F, Dieterich C (2016) Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 32:1094–1096
Hansen TB, Venø MT, Damgaard CK, Kjems J (2016) Comparison of circular RNA prediction tools. Nucleic Acids Res 44:e58
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 2016. https://doi.org/10.1101/gr.202895.115
Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16:4
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20:1666–1670
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13:34–42
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42(Database issue):D92–D97
Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12:R72
Hansen TB (2018) Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol 6:20
Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126
Izuogu OG, Alhasan AA, Alafghani HM, Santibanez-Koref M, Elliott DJ, Jackson MS (2016) PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. BMC Bioinformatics 17:31
Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY (2016) NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res 44:e29
Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S et al (2014) A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol 15:R34
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K et al (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44:e87
Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL et al (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38:e178
Acknowledgments
This work was supported in full by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply
About this protocol
Cite this protocol
Cochran, K.R., Gorospe, M., De, S. (2022). Bioinformatic Analysis of CircRNA from RNA-seq Datasets. In: Cortassa, S., Aon, M.A. (eds) Computational Systems Biology in Medicine and Biotechnology. Methods in Molecular Biology, vol 2399. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1831-8_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1831-8_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1830-1
Online ISBN: 978-1-0716-1831-8
eBook Packages: Springer Protocols