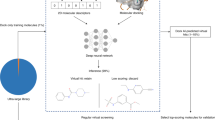
Abstract
Ultrahigh-throughput virtual screening (uHTVS) is an emerging field linking together classical docking techniques with high-throughput AI methods. We outline mechanistic docking models’ goals and successes. We present different AI accelerated workflows for uHTVS, mainly through surrogate docking models. We showcase a novel feature representation technique, molecular depictions (images), as a surrogate model for docking. Along with a discussion on analyzing screens using regression enrichment surfaces at the tens of billion scale, we outline a future for uHTVS screening pipelines with deep learning.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Rester U (2008) From virtuality to reality-Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Devel 11:559
Ltd E Enamine REAL Space
Lahue BR, Glick M, Tudor M et al (2020) Diversity & tractability revisited in collaborative small molecule phenotypic screening library design. Bioorg Med Chem 28:115192
Paricharak S, Méndez-Lucio O, Chavan Ravindranath A et al (2018) Data-driven approaches used for compound library design, hit triage and bioactivity modeling in high-throughput screening. Brief Bioinform 19:277–285
Lyu J, Wang S, Balius TE et al (2019) Ultra-large library docking for discovering new chemotypes. Nature 566:224–229
Jia X, Lynch A, Huang Y et al (2019) Anthropogenic biases in chemical reaction data hinder exploratory inorganic synthesis. Nature 573:251–255
Su AI, Lorber DM, Weston GS et al (2001) Docking molecules by families to increase the diversity of hits in database screens: computational strategy and experimental evaluation. Proteins 42:279–293
Polishchuk PG, Madzhidov TI, Varnek A (2013) Estimation of the size of drug-like chemical space based on GDB-17 data. J Comput Aided Mol Des 27:675–679
Bolte M, Hogan CJ (1995) Conflict over the age of the Universe. Nature 376:399–402
Schneider G (2010) Virtual screening: an endless staircase? Nat Rev Drug Discov 9:273–276
McInnes C (2007) Virtual screening strategies in drug discovery. Curr Opin Chem Biol 11:494–502
Sliwoski G, Kothiwale S, Meiler J, Lowe EW Jr (2014) Computational methods in drug discovery. Pharmacol Rev 66:334–395. https://doi.org/10.1124/pr.112.007336
Sakkiah S, Thangapandian S, John S et al (2010) 3D QSAR pharmacophore based virtual screening and molecular docking for identification of potential HSP90 inhibitors. Eur J Med Chem 45:2132–2140
Sun H (2008) Pharmacophore-based virtual screening. Curr Med Chem 15:1018–1024
Willett P, Barnard JM, Downs GM (1998) Chemical similarity searching. J Chem Inf Comput Sci 38:983–996
Kumar A, Zhang KY (2018) Advances in the development of shape similarity methods and their application in drug discovery. Front Chem 6:315
Coley CW, Barzilay R, Green WH et al (2017) Convolutional embedding of attributed molecular graphs for physical property prediction. J Chem Inf Model 57:1757–1772
Liu Z, Du J, Fang J, et al (2019) DeepScreening: a deep learning-based screening web server for accelerating drug discovery Database 2019
Zhou H, Skolnick J (2013) FINDSITEcomb: a threading/structure-based, proteomic-scale virtual ligand screening approach. J Chem Inf Model 53:230–240
Oprea TI (2000) Current trends in lead discovery: are we looking for the appropriate properties? Mol Divers 5:199–208
Verdonk ML, Berdini V, Hartshorn MJ et al (2004) Virtual screening using protein- ligand docking: avoiding artificial enrichment. J Chem Inf Comput Sci 44:793–806
Klebe G (2006) Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today 11:580–594. https://doi.org/10.1016/j.drudis.2006.05.012
Sterling T, Irwin JJ (2015) ZINC 15-ligand discovery for everyone. J Chem Inf Model 55:2324–2337
Shivanyuk A, Ryabukhin S, Tolmachev A et al (2007) Enamine real database: making chemical diversity real. Chem Today 25:58–59
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: An open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
Le Guilloux V, Schmidtke P, Tuffery P (2009) Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10:1–11
Bernstein FC, Koetzle TF, Williams GJ et al (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. Eur J Biochem 80:319–324
Warren GL, Andrews CW, Capelli A-M et al (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931
Cole JC, Murray CW, Nissink JWM et al (2005) Comparing protein–ligand docking programs is difficult. Proteins 60:325–332
Kitchen D, Decornez H, Furr J, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949. https://doi.org/10.1038/nrd1549
Ballester PJ, Mitchell JB (2010) A machine learning approach to predicting protein–ligand binding affinity with applications to molecular docking. Bioinformatics 26:1169–1175
Mcgann MR, Almond HR, Nicholls A et al (2003) Gaussian docking functions. Biopolymers 68:76–90
Guedes IA, Pereira FS, Dardenne LE (2018) Empirical scoring functions for structure-based virtual screening: applications, critical aspects, and challenges. Front Pharmacol 9:1089
Clark RD, Strizhev A, Leonard JM et al (2002) Consensus scoring for ligand/protein interactions. J Mol Graph Model 20:281–295
Meiler J, Baker D (2006) ROSETTALIGAND: Protein–small molecule docking with full side-chain flexibility. Proteins 65:538–548
Razzaghi-Asl N, Sepehri S, Ebadi A et al (2015) Effect of biomolecular conformation on docking simulation: a case study on a potent HIV-1 protease inhibitor. Iran J Pharm Res 14:785
McGaughey GB, Sheridan RP, Bayly CI et al (2007) Comparison of topological, shape, and docking methods in virtual screening. J Chem Inf Model 47:1504–1519
Francoeur PG, Masuda T, Sunseri J et al (2020) Three-dimensional convolutional neural networks and a cross-docked data set for structure-based drug design. J Chem Inf Model 60:4200–4215. https://doi.org/10.1021/acs.jcim.0c00411
Sunseri J, King JE, Francoeur PG, Koes DR (2019) Convolutional neural network scoring and minimization in the D3R 2017 community challenge. J Comput Aided Mol Des 33:19–34. https://doi.org/10.1007/s10822-018-0133-y
Xu Z, Wauchope OR, Frank AT (2020) Navigating chemical space by interfacing generative artificial intelligence and molecular docking. bioRxiv
Li X, Xu Y, Yao H, Lin K (2020) Chemical space exploration based on recurrent neural networks: applications in discovering kinase inhibitors. J Chem 12:1–13
Landrum G et al (2006) RDKit: open-source cheminformatics
Pechan I, Feher B (2011) Molecular docking on FPGA and GPU platforms. In: 2011 21st international conference on field programmable logic and applications. IEEE, pp 474–477
LeGrand S, Scheinberg A, Tillack AF, et al (2020) GPU-accelerated drug discovery with docking on the summit supercomputer: porting, optimization, and application to COVID-19 research. In: Proceedings of the 11th ACM international conference on bioinformatics, computational biology and health informatics, pp 1–10
Zlateski A, Lee K, Seung HS (2016) ZNNi: maximizing the inference throughput of 3D convolutional networks on CPUs and GPUs. In: SC’16: Proceedings of the international conference for high performance computing, networking, storage and analysis. IEEE, pp 854–865
Lee H, Merzky A, Tan L, et al (2020) Scalable HPC and AI infrastructure for COVID-19 therapeutics. arXiv preprint arXiv:201010517
Wright D, Devitt-Lee A, Clyde A, et al (2019) Combining molecular simulation and machine learning to INSPIRE improved cancer therapy. In: CompBioMed conference 2019
Lu S-Y, Jiang Y-J, Lv J et al (2010) Molecular docking and molecular dynamics simulation studies of GPR40 receptor–agonist interactions. J Mol Graph Model 28:766–774
Schütt KT, Sauceda HE, Kindermans P-J et al (2018) SchNet—a deep learning architecture for molecules and materials. J Chem Phys 148:241722
Bartók AP, De S, Poelking C et al (2017) Machine learning unifies the modeling of materials and molecules. Sci Adv 3:e1701816
Pastor M, Cruciani G, McLay I et al (2000) GRid-INdependent descriptors (GRIND): a novel class of alignment-independent three-dimensional molecular descriptors. J Med Chem 43:3233–3243
Yap CW (2011) PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1474
Todeschini R, Consonni V (2008) Handbook of molecular descriptors. John Wiley & Sons, Hoboken, NJ
Moriwaki H, Tian Y-S, Kawashita N, Takagi T (2018) Mordred: a molecular descriptor calculator. J Chem 10:4
Clark AM, Labute P, Santavy M (2006) 2D structure depiction. J Chem Inf Model 46:1107–1123
Ebalunode JO, Zheng W (2009) Unconventional 2D shape similarity method affords comparable enrichment as a 3D shape method in virtual screening experiments. J Chem Inf Model 49:1313–1320
Babel O (2010) The open source chemistry toolbox
OEChem T (2012) OpenEye Scientific Software. Inc, Santa Fe, NM, USA
Duvenaud DK, Maclaurin D, Iparraguirre J, et al (2015) Convolutional networks on graphs for learning molecular fingerprints. In: Advances in neural information processing systems. The Neural Information Processing Systems Foundation. pp. 2224–2232
Zhou J, Cui G, Zhang Z, et al (2018) Graph neural networks: a review of methods and applications. arXiv preprint arXiv:181208434
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. preprint arXiv:1609.02907
Elton DC, Boukouvalas Z, Fuge MD, Chung PW (2019) Deep learning for molecular design—a review of the state of the art. Mol Syst Des Eng 4:828–849
Tripathi A, Bankaitis VA (2017) Molecular docking: From lock and key to combination lock. J Mol Med Clin Appl 2
Deng J, Dong W, Socher R, et al (2009) Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, pp 248–255
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Nam H, Ha J-W, Kim J (2017) Dual attention networks for multimodal reasoning and matching. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 299–307
Sundararajan M, Taly A, Yan Q (2017) Axiomatic attribution for deep networks. arXiv preprint arXiv:170301365
Raghuraman A, Mosier PD, Desai UR (2006) Finding a needle in a haystack: development of a combinatorial virtual screening approach for identifying high specificity heparin/heparan sulfate sequence (s). J Med Chem 49:3553–3562
Da C, Stashko M, Jayakody C et al (2015) Discovery of Mer kinase inhibitors by virtual screening using structural protein–ligand interaction fingerprints. Bioorg Med Chem 23:1096–1101
Cheong R, Wang CJ, Levchenko A (2009) High content cell screening in a microfluidic device. Mol Cell Proteomics 8:433–442
Feinberg EN, Sur D, Wu Z et al (2018) PotentialNet for molecular property prediction. ACS Centr Sci 4:1520–1530
Irwin JJ, Shoichet BK, Mysinger MM et al (2009) Automated docking screens: a feasibility study. J Med Chem 52:5712–5720
Malo N, Hanley JA, Cerquozzi S et al (2006) Statistical practice in high-throughput screening data analysis. Nat Biotechnol 24:167–175
Clyde A, Duan X, Stevens R (2020) Regression enrichment surfaces: a simple analysis technique for virtual drug screening models. arXiv preprint arXiv:200601171
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Clyde, A. (2022). Ultrahigh Throughput Protein–Ligand Docking with Deep Learning. In: Heifetz, A. (eds) Artificial Intelligence in Drug Design. Methods in Molecular Biology, vol 2390. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1787-8_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1787-8_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1786-1
Online ISBN: 978-1-0716-1787-8
eBook Packages: Springer Protocols