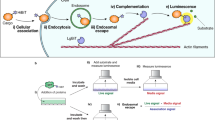
Abstract
Target engagement and cell permeation are important parameters that may limit the efficacy of proteolysis-targeting chimeras (PROTACs). Here, we present an approach that facilitates both the quantitation of PROTAC binding affinity for an E3 ligase of interest, as well as the assessment of relative intracellular availability. We present a panel of E3 ligase target engagement assays based upon the NanoBRET Target Engagement platform. Querying E3 ligase engagement under live-cell and permeabilized-cell conditions allow calculation of an availability index that can be used to rank order the intracellular availability of PROTACs. Here we present examples where the cellular availability of PROTACs and their monovalent precursors are prioritized using NanoBRET assays for CRBN or VHL E3 ligases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Shapira M, Calabrese MF, Bullock AN, Crews CM (2019) Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 18(12):949–963. https://doi.org/10.1038/s41573-019-0047-y
Daniels DL, Riching KM, Urh M (2019) Monitoring and deciphering protein degradation pathways inside cells. Drug Discov Today Technol 31:61–68. https://doi.org/10.1016/j.ddtec.2018.12.001
Liu J, Ma J, Xia J, Li Y, Wang ZP, Wei W (2020) PROTACs: a novel strategy for cancer therapy. Semin Cancer Biol 67(Pt 2):171–179. https://doi.org/10.1016/j.semcancer.2020.02.006
Maple HJ, Clayden N, Baron A, Stacey C, Felix R (2019) Developing degraders: principles and perspectives on design and chemical space. MedChemComm 10(10):1755–1764. https://doi.org/10.1039/c9md00272c
Soares P, Gadd MS, Frost J, Galdeano C, Ellis L, Epemolu O, Rocha S, Read KD, Ciulli A (2018) Group-Based Optimization of Potent and Cell-Active Inhibitors of the Von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298). J Med Chem 61(2):599–618. https://doi.org/10.1021/acs.jmedchem.7b00675
Foley CA, Potjewyd F, Lamb KN, James LI, Frye SV (2020) Assessing the cell permeability of bivalent chemical degraders using the chloroalkane penetration assay. ACS Chem Biol 15(1):290–295
Zoppi V, Hughes SJ, Maniaci C, Testa A, Gmaschitz T, Wieshofer C, Koegl M, Riching KM, Daniels DL, Spallarossa A, Ciulli A (2019) Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von hipper-Lindau (VHL) based dual degrader probe of BRD9 and BRD7. J Med Chem 62(2):699–726
Robers MB, Dart ML, Woodroofe CC, Zimprich CA, Kirkland TA, Machleidt T, Kupcho KR, Levin S, Hartnett JR, Zimmerman K, Niles AL, Ohana RF, Daniels DL, Slater M, Wood MG, Cong M, Cheng YQ, Wood KV (2015) Target engagement and drug residence time can be observed in living cells with BRET. Nat Commun 6:10091. https://doi.org/10.1038/ncomms10091
Robers MB, Vasta JD, Corona CR, Ohana RF, Hurst R, Jhala MA, Comess KM, Wood KV (2019) Quantitative, real-time measurements of intracellular target engagement using energy transfer. Methods Mol Biol 1888:45–71. https://doi.org/10.1007/978-1-4939-8891-4_3
Vasta JD, Corona CR, Wilkinson J, Zimprich CA, Hartnett JR, Ingold MR, Zimmerman K, Machleidt T, Kirkland TA, Huwiler KG, Ohana RF, Slater M, Otto P, Cong M, Wells CI, Berger BT, Hanke T, Glas C, Ding K, Drewry DH, Huber KVM, Willson TM, Knapp S, Muller S, Meisenheimer PL, Fan F, Wood KV, Robers MB (2018) Quantitative, wide-Spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem Biol 25(2):206–214. e211. https://doi.org/10.1016/j.chembiol.2017.10.010
Mateus A, Treyer A, Wegler C, Karlgren M, Matsson P, Artursson P (2017) Intracellular drug bioavailability: a new predictor of system-dependent drug disposition. Sci Rep 7:43047. https://doi.org/10.1038/srep43047
Hulme EC, Trevethick MA (2010) Ligand binding assays at equilbrium: validation and interpretation. Br J Pharmacol 161:1219–1237
Frost J, Galdeano C, Soares P, Gadd MS, Grzes KM, Ellis L, Epemolu O, Shimamura S, Bantscheff M, Grandi P, Read KD, Cantrell DA, Rocha S, Ciulli A (2016) Potent and selective chemical probe of hypoxic signaling downstream of HIF-α hydroxylation via VHL inhibition. Nat Commun 7:13312. https://doi.org/10.1038/ncomms13312
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AMK, Wang J, Chen X, Dong H, Siu K, Winkler JD, Crew AP, Crews CM, Coleman KG (2016) PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 113(26):7124–7129. https://doi.org/10.1073/pnas.1521738113
Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, Iulio JD, Souza A, Ott CJ, Roberts JM, Zeid R, Scott TG, Paulk J, Lachance K, Olson CM, Dastjerdi S, Bauer S, Lin CY, Gray NS, Kelliher MA, Churchman LS, Bradner JE (2017) BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell 67(1):5–18. https://doi.org/10.1016/j.molcel.2017.06.004
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Vasta, J.D., Corona, C.R., Robers, M.B. (2021). A High-Throughput Method to Prioritize PROTAC Intracellular Target Engagement and Cell Permeability Using NanoBRET. In: Cacace, A.M., Hickey, C.M., Békés, M. (eds) Targeted Protein Degradation. Methods in Molecular Biology, vol 2365. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1665-9_14
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1665-9_14
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1664-2
Online ISBN: 978-1-0716-1665-9
eBook Packages: Springer Protocols