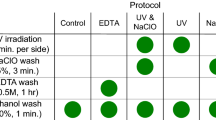
Abstract
Ancient dental calculus provides a challenging, yet unparalleled, opportunity to reconstruct ancient oral microbial communities and trace the origins of modern microbiota-associated diseases. Metagenomic analysis of ancient dental calculus using high-throughput DNA sequencing has proven itself as an effective method to accurately reconstruct microorganisms that once lived in the mouths of ancient humans. Here, we provide the strategy, methodologies, and approaches used to establish an ancient dental calculus project, from project conception, community engagement, sampling, extracting DNA, and preparing shotgun metagenomic DNA libraries for sequencing on an Illumina platform. We also discuss techniques to minimize background or contaminant DNA by monitoring and reducing contamination in calculus data sets, utilizing appropriate protective gear, and employing the use of sample decontamination strategies. In this methodology chapter, we hope to promote transparency in the ancient dental calculus research field and encourage collaboration across the ancient DNA research community.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M et al (2010) A draft sequence of the Neandertal genome. Science 328:710–722. https://doi.org/10.1126/science.1188021
Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N et al (2004) Genetic analyses from ancient DNA. Annu Rev Genet 38:645–679. https://doi.org/10.1146/annurev.genet.37.110801.143214
Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML et al (2012) The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc Biol Sci 279:4724–4733. https://doi.org/10.1098/rspb.2012.1745
Turney CSM, Fogwill CJ, Golledge NR, McKay NP, van Sebille E, Jones RT et al (2020) Early last interglacial ocean warming drove substantial ice mass loss from Antarctica. Proc Natl Acad Sci U S A 117:3996. https://doi.org/10.1073/pnas.1902469117
Frisia S, Weyrich LS, Hellstrom J, Borsato A, Golledge NR, Anesio AM et al (2017) The influence of Antarctic subglacial volcanism on the global iron cycle during the last glacial Mmaximum. Nat Commun 8:15425. https://doi.org/10.1038/ncomms15425
Zobel M, Davison J, Edwards ME, Brochmann C, Coissac E, Taberlet P et al (2018) Ancient environmental DNA reveals shifts in dominant mutualisms during the late quaternary. Nat Commun 9:139. https://doi.org/10.1038/s41467-017-02421-3
Boast AP, Weyrich LS, Wood JR, Metcalf JL, Knight R, Cooper A (2018) Coprolites reveal ecological interactions lost with the extinction of New Zealand birds. Proc Natl Acad Sci U S A 115:1546. https://doi.org/10.1073/pnas.1712337115
Weyrich LS, Duchene S, Soubrier J, Arriola L, Llamas B, Breen J et al (2017) Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544:357–361. https://doi.org/10.1038/nature21674
Warinner C, Speller C, Collins MJ, Lewis CM (2015) Ancient human microbiomes. J Hum Evol 79:125–136. https://doi.org/10.1016/j.jhevol.2014.10.016
Weyrich LS, Dobney K, Cooper A (2015) Ancient DNA analysis of dental calculus. J Hum Evol 79:119–124. https://doi.org/10.1016/j.jhevol.2014.06.018
Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W et al (2013) Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and industrial revolutions. Nat Genet 45:450–455. https://doi.org/10.1038/ng.2536
Warinner C, Rodrigues JFM, Vyas R, Trachsel C, Shved N, Grossmann J et al (2014) Pathogens and host immunity in the ancient human oral cavity. Nat Genet 46:336–344. https://doi.org/10.1038/ng.2906
Farrer AG, Bekvalac J, Redfern R, Gully N, Dobney K, Cooper A et al (2018) Biological and cultural drivers of oral microbiota in Medieval and Post-Medieval London, UK. bioRxiv 343889. https://doi.org/10.1101/343889
Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L (2013) mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29:1682–1684. https://doi.org/10.1093/bioinformatics/btt193
Warinner C, Herbig A, Mann A, Fellows Yates JA, Weiß CL, Burbano HA et al (2017) A robust framework for microbial archaeology. Annu Rev Genomics Hum Genet 18:321–356. https://doi.org/10.1146/annurev-genom-091416-035526
Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, Weyrich LS (2019) Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol 27:105–117. https://doi.org/10.1016/j.tim.2018.11.003
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF et al (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. https://doi.org/10.1186/s12915-014-0087-z
Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, Najar F et al (2012) Insights from characterizing extinct human gut microbiomes. PLoS One 7:e51146. https://doi.org/10.1371/journal.pone.0051146
Austin RM, Sholts SB, Williams L, Kistler L, Hofman CA (2019) Opinion: to curate the molecular past, museums need a carefully considered set of best practices. Proc Natl Acad Sci U S A 116:1471. https://doi.org/10.1073/pnas.1822038116
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Ziesemer KA, Mann AE, Sankaranarayanan K, Schroeder H, Ozga AT, Brandt BW et al (2015) Intrinsic challenges in ancient microbiome reconstruction using 16S rRNA gene amplification. Sci Rep 5:16498. https://doi.org/10.1038/srep16498
Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B et al (2013) Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A 110:15758. https://doi.org/10.1073/pnas.1314445110
Wagner JK, Colwell C, Claw KG, Stone AC, Bolnick DA, Hawks J et al (2020) Fostering responsible research on ancient DNA. Am J Hum Genet 107:183–195. https://doi.org/10.1016/j.ajhg.2020.06.017
Fagernäs Z, García-Collado MI, Hendy J, Hofman CA, Speller C, Velsko I et al (2020) A unified protocol for simultaneous extraction of DNA and proteins from archaeological dental calculus. J Archaeol Sci 118:105135. https://doi.org/10.1016/j.jas.2020.105135
Kircher M, Sawyer S, Meyer M (2012) Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. https://doi.org/10.1093/nar/gkr771
Llamas B, Valverde G, Fehren-Schmitz L, Weyrich LS, Cooper A, Haak W et al (2016) From the field to the laboratory: controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. Sci Technol Archeol Res 3:1–14. https://doi.org/10.1080/20548923.2016.1258824
Farrer AG, Wright SL, Skelly F et al (2021) Effectiveness of decontamination protocols when analyzing ancient DNA preserved in dental calculus. Sci Rep 11, 7456. https://doi.org/10.1038/s41598-021-86100-w
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Moore, N.E., Weyrich, L.S. (2021). A Standardized Approach for Shotgun Metagenomic Analysis of Ancient Dental Calculus . In: Adami, G.R. (eds) The Oral Microbiome. Methods in Molecular Biology, vol 2327. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1518-8_7
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1518-8_7
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1517-1
Online ISBN: 978-1-0716-1518-8
eBook Packages: Springer Protocols