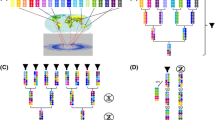
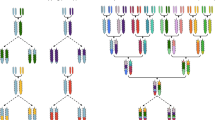
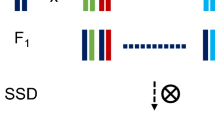
Abstract
Multiparental populations are located midway between association mapping that relies on germplasm collections and classic linkage analysis, based upon biparental populations. They provide several key advantages such as the possibility to include a higher number of alleles and increased level of recombination with respect to biparental populations, and more equilibrated allelic frequencies than association mapping panels. Moreover, in these populations new allele’s combinations arise from recombination that may reveal transgressive phenotypes and make them a useful pre-breeding material. Here we describe the strategies for working with multiparental populations, focusing on nested association mapping populations (NAM) and multiparent advanced generation intercross populations (MAGIC). We provide details from the selection of founders, population development, and characterization to the statistical methods for genetic mapping and quantitative trait detection.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Morrell PL, Buckler ES, Ross-Ibarra J (2012) Crop genomics: advances and applications. Nat Rev Genet 13:85–96
Price AH (2006) Believe it or not, QTLs are accurate! Trends Plant Sci 11:213–216
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9:29
Visscher PM, Brown MA, McCarthy MI, Yang J (2012) Five years of GWAS discovery. Am J Hum Genet 90:7–24
Churchill G, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J et al (2004) The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36:1133–1137
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551
Mackay I, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends Plant Sci 12:57–63
Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol 11:215–221
Huang BE, Verbyla KL, Verbyla AP, Raghavan C, Singh VK, Gaur P et al (2015) MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128:999–1017
Pascual L, Albert E, Sauvage C, Duangjit J, Bouchet J-P, Bitton F et al (2016) Dissecting quantitative trait variation in the resequencing era: complementarity of bi-parental, multi-parental and association panels. Plant Sci 242:120–130
Stich B (2009) Comparison of mating designs for establishing nested association mapping populations in maize and Arabidopsis thaliana. Genetics 183:1525–1534
Bauer E, Falque M, Walter H et al (2013) Intraspecific variation of recombination rate in maize. Genome Biol 14:R103
Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E et al (2015) Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16(1):290
Nice LM, Steffenson BJ, Brown-Guedira GL, Akhunov ED, Liu C, Kono TJY et al (2016) Development and genetic characterization of an advanced backcross-nested association mapping (AB-NAM) population of wild × cultivated barley. Genetics 203:1453
Bajgain P, Rouse MN, Tsilo TJ, Macharia GK, Bhavani S, Jin Y et al (2016) Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PLoS One 11:e0155760
Fragoso CA, Moreno M, Wang Z, Heffelfinger C, Arbelaez LJ, Aguirre JA et al (2017) Genetic architecture of a rice nested association mapping population. G3 7:1913–1926
Bouchet S, Olatoye MO, Marla SR, Perumal R, Tesso T, Yu J et al (2017) Increased power to dissect adaptive traits in global Sorghum diversity using a nested association mapping population. Genetics 206:573–585
Hu J, Guo C, Wang B, Ye J, Liu M, Wu Z et al (2018) Genetic properties of a nested association mapping population constructed with semi-winter and spring oilseed rapes. Front Plant Sci 9:1740
Diers BW, Specht J, Rainey KM, Cregan P, Song Q, Ramasubramanian V et al (2018) Genetic architecture of soybean yield and agronomic traits. G3 8:3367–3375
Jordan KW, Wang S, He F, Chao S, Lun Y, Paux E et al (2018) The genetic architecture of genome-wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J 95:1039–1054
Chen Q, Yang CJ, York AM, Xue W, Daskalska LL, DeValk CA et al (2019) TeoNAM: a nested association mapping population for domestication and agronomic trait analysis in maize. Genetics 213:1065–1078
Hemshrot A, Poets AM, Tyagi P, Lei L, Carter CK, Hirsch CN et al (2019) Development of a multiparent population for genetic mapping and allele discovery in six-row barley. Genetics 213:595–613
Marla SR, Burow G, Chopra R, Hayes C, Olatoye MO, Felderhoff T et al (2019) Genetic architecture of chilling tolerance in Sorghum dissected with a nested association mapping population. G3 9:4045–4057
Kidane YG, Gesesse CA, Hailemariam BN, Desta EA, Mengistu DK, Fadda C et al (2019) A large nested association mapping population for breeding and quantitative trait locus mapping in Ethiopian durum wheat. Plant Biotechnol J 17:1380–1393
Thachuk C, Crossa J, Franco J, Dreisigacker S, Warburton M, Davenport GF (2009) Core Hunter: an algorithm for sampling genetic resources based on multiple genetic measure. BMC Bioinformatics 10:243
Knott DR, Kumar J (1975) Comparison of early generation yield testing and a single seed descent procedure in wheat breeding. Crop Sci 15:295–299
Guo B, Sleper DA, Beavis WD (2010) Nested association mapping for identification of functional markers. Genetics 186:373–383
Klasen JR, Piepho HP, Stich B (2012) QTL detection power of multi-parental RIL populations in Arabidopsis thaliana. Heredity 108:626–632
Li J, Bus A, Spamer V, Stich B (2016) Comparison of statistical models for nested association mapping in rapeseed (Brassica napus eL.) through computer simulations. BMC Plant Biol 16:26
Griffing B (1956) Concept of general and specific combining ability in relation to diallel crossing systems. Aust J Biol Sci 9:463–493
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253
Sansaloni C, Petroli C, Jaccoud D et al (2011) Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc 5:P54
McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q et al (2009) Genetic properties of the maize nested association mapping population. Science 325:737–740
Guo B, Beavis WD (2011) In silico genotyping of the maize nested association mapping population. Mol Breed 27:107–113
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e1937
Zan Y, Payen T, Lillie M, Honaker CF, Siegel PB, Carlborg O (2019) Genotyping by low-coverage whole-genome sequencing in intercross pedigrees from outbred founders: a cost-efficient approach. Genet Select Evol 51:44
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Islam MS, Thyssen GN, Jenkins JN, Zeng L, Delhom CD, McCarty JC et al (2016) A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genomics 17. https://doi.org/10.1186/s12864-016-3249-2
Bandillo N, Raghavan C, Muyco PA, Sevilla MAL, Lobina IT, Dilla-Ermita CJ et al (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6:11
Ongom PO, Ejeta G (2018) Mating design and genetic structure of a multi-parent advanced generation intercross (MAGIC) population of Sorghum (Sorghum bicolor (L.) Moench). G3 8:331–341
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ et al (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Naoumkina M, Thyssen GN, Fang DD, Jenkins JN, McCarty JC, Florane CB (2019) Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L) MAGIC population. BMC Genomics 20:112
Butler D, Cullis B, Gilmour A, Gogel B (2007) ASRemlR reference manual. State of Queensland Department of Primary Industries and Fisheries
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P et al (2017) Linkage analysis and association mapping QTL detection models for hybrids between multiparental population from two heterotic groups: application to biomass production in maize (Zea mays L.). G3 7:3649–3657
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P et al (2017) Reciprocal genetics: identifying QTL for general and specific combining abilities in hybrids between multiparental populations from two maize (Zea mays L.) heterotic groups. Genetics 207:1167–1180
Mott R, Talbot CJ, Turri MG, Collins AC, Flint J (2000) A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A 97:12649–12654
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK et al (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10:826–839
Gnan S, Priest A, Kover PX (2014) The genetic basis of natural variation in seed size and seed number and their trade-off using Arabidopsis thaliana MAGIC lines. Genetics 198:1751
Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD et al (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5:e1000551
Huang BE, George AW (2011) R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27:727–729
Sannemann W, Huang BE, Mathew B, Leon J (2015) Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol Breed 35:86
Stadlmeier M, Hartl L, Mohler V (2018) Usefulness of a multiparent advanced generation intercross population with a greatly reduced mating design for genetic studies in winter wheat. Front Plant Sci 9:1825
Pascual L, Desplat N, Huang BE, Desgroux A, Bruguier L, Bouchet J-P et al (2015) Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol J 13:565–577
Huynh B-L, Ehlers JD, Huang BE, Munoz-Amatriain M, Lonardi S, Santos JRP et al (2018) A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J 93:1129–1142
Broman KW, Gatti DM, Simecek P, Furlotte NA, Prins P, Sen S et al (2019) R/qtl2: software for mapping quantitative trait loci with high-dimensional data and multiparent populations. Genetics 211:495–502
de Jong M, Tavares H, Pasam RK, Butler R, Ward S, George G et al (2019) Natural variation in Arabidopsis shoot branching plasticity in response to nitrate supply affects fitness. PLoS Genet 15:e100836
Wei J, Xu S (2016) A random-model approach to QTL mapping in multiparent advanced generation intercross (MAGIC) populations. Genetics 202:471
Verbyla AP, George AW, Cavanagh CR, Verbyla KL (2014) Whole-genome QTL analysis for MAGIC. Theor Appl Genet 127:1753–1770
Verbyla AP, Cavanagh CR, Verbyla KL (2014) Whole-genome analysis of multienvironment or multitrait QTL in MAGIC. G3 4:1569–1584
Zhang L, Meng L, Wang J (2019) Linkage analysis and integrated software GAPL for pure-line populations derived from four-way and eight-way crosses. Crop J 7:283–293
Shi J, Wang J, Zhang L (2019) Genetic mapping with background control for quantitative trait locus (QTL) in 8-parental pure-line populations. J Hered 110:880–891
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Butron A, Santiago R, Cao A, Samayoa LF, Malvar RA (2019) QTLs for resistance to Fusarium ear rot in a multiparent advanced generation intercross (MAGIC) maize population. Plant Dis 103:897–904
Mackay IJ, Bansept-Basler P, Barber T, Bentley AR, Cockram J, Gosman N et al (2014) An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 4:1603–1610
Sallam A, Martsch R (2015) Association mapping for frost tolerance using multi-parent advanced generation inter-cross (MAGIC) population in faba bean (Vicia faba L.). Genetica 143:501–514
Campanelli G, Sestili S, Acciarri N, Montemurro F, Palma D, Leteo F et al (2019) Multi-parental advances generation inter-cross population, to develop organic tomato genotypes by participatory plant breeding. Agronomy 9:119
Meng L, Zhao X, Ponce K, Ye G, Leung H (2016) QTL mapping for agronomic traits using multi-parent advanced generation inter-cross (MAGIC) populations derived from diverse elite indica rice lines. Field Crops Res 189:19–42
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351
Diouf IA, Derivot L, Bitton F, Pascual L, Causse M (2018) Water deficit and salinity stress reveal many specific QTL for plant growth and fruit quality traits in tomato. Front Plant Sci 9:279
Ponce KS, Ye G, Zhao X (2018) QTL identification for cooking and eating quality in indica rice using multi-parent advanced generation intercross (MAGIC) population. Front Plant Sci 9:868
Valente F, Gauthier F, Bardol N, Blanc G, Joets J, Charcosset A et al (2014) OptiMAS: a decision support tool to conduct marker-assisted selection programs. Crop Breed Methods Protoc 1145:97–116
R Core Team (2019) R: a language and environment for statistical computing. In: R Foundation for Statistical Computing. Available via DIALOG. https://www.R-project.org/. Accessed 31 Jan 2020
Gardner KA, Wittern LM, Mackay IJ (2016) A highly recombined, high-density, eight-founder wheat MAGIC map reveals extensive segregation distortion and genomic locations of introgression segments. Plant Biotechnol J 14:1406–1417
Shah R, Huang E (2019) Map construction using multi-parent populations (Version v0.0.6). In: Zenodo. Available via DIALOG. https://doi.org/10.5281/zenodo.2613114. Accessed 31 Jan 2020
Zheng C, Boer MP, van Eeuwijk FA (2019) Construction of genetic linkage maps in multiparental populations. Genetics 212:1031–1044
Ogawa D, Yamamoto E, Ohtani T, Kanno N, Tsunematsu H, Nonoue Y et al (2018) Haplotype-based allele mining in the Japan-MAGIC rice population. Sci Rep 8:4379
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Li H, Bradbury P, Ersoz E, Buckler ES, Wang J (2011) Joint QTL linkage mapping for multiple-cross mating design sharing one common parent. PLoS One 6:e17573
Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-Y, Freimer NB et al (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42:348–U110
Xavier A, Xu S, Muir WM, Rainey KM (2015) NAM: association studies in multiple populations. Bioinformatics 31:3862–3864
Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C et al (2009) The genetic architecture of maize flowering time. Science 325:714–718
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
SAS Institute (2011) SAS/STAT 9.3 user’s guide. SAS Institute Inc, Cary, NC, USA. Available via DIALOG. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.226.6407&rep=rep1&type=pdf. Accessed 31 Jan 2020
Bian Y, Holland JB (2015) Ensemble learning of QTL models improves prediction of complex traits. G3 5:2073–2084
Dietterich TG (2000) Ensemble methods in machine learning. In: International workshop on multiple classifier systems. Springer, Berlin, pp 1–15
Lehermeier C, Kramer N, Bauer E et al (2014) Usefulness of multiparental populations of maize (Zea mays L.) for genome-based prediction. Genetics 198:3–16
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Diouf, I., Pascual, L. (2021). Multiparental Population in Crops: Methods of Development and Dissection of Genetic Traits. In: Tripodi, P. (eds) Crop Breeding. Methods in Molecular Biology, vol 2264. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1201-9_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1201-9_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1200-2
Online ISBN: 978-1-0716-1201-9
eBook Packages: Springer Protocols