Abstract
Copy number variants (CNVs) refer to the loss or gain of copies of a genomic DNA region. While some CNVs may play a role in species evolution by enriching the diversity of an organism, CNVs may also be linked to certain diseases such as neurological disorders, early onset obesity, and cancer. CNVs may affect gene expression by direct overlap of the genic region or by an indirect effect where the CNV is located outside the gene location. These indirect CNV regions may contain regulatory elements such as transcription enhancers or repressors as well as regulators such as miRNAs which may work at the level of transcription or translation. Danio rerio (zebrafish) is an excellent model organism for CNV studies. Zebrafish genomes contain a large amount of variation with 14.6% of the zebrafish reference genome found to be copy number variable. This level of variation is more than four times the percentage of reference genome sequence covered by similarly common CNVs in humans. It is this high level of variation that makes zebrafish interesting to investigate the effects of CNV on gene expression. Additionally, zebrafish share 70% of genetic similarities with humans, and 84% of genes associated with human disease are also found in zebrafish. Expressive quantitative trait loci (eQTL) analysis may be used in zebrafish to explore how CNVs may affect gene expression in both a direct and indirect manner. eQTL analysis may be performed for cis associations with a 1-Mb (megabase) window upstream and downstream from the transcription probe midpoint to CGH midpoint. Trans associations (variants that are located beyond the 1-Mb window of the gene either on the same chromosome as the gene or on a different chromosome) may be investigated as well through eQTL analysis; however, trans associations require more tests to be performed than cis associations, which limits power to detect associations. Pairwise associations between each pair of copy number variant and gene will be investigated separately from the same individual using Spearman rank correlations with significant associations found being followed with a multi-test correction technique to assess significance of those CNV gene expression associations. An association between a CNV to a gene expression phenotype should be considered significant only if the p value from the analysis of the observed data is lower than the 0.001 tail threshold from a distribution of the minimal p values (which are found from all comparisons for a given gene from 10,000 permutations of the expression phenotypes). Associations between CNVs and genes may be found to be direct or indirect as well as positive (increased copy number—increased expression) or negative (increased copy number—decreased expression, decreased copy number—increased expression). Ongoing analyses with these associations will investigate the impact of CNVs on gene functionality including immune function and potential disease susceptibility.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Beckmann JS, Estivill X, Antonarakis SE (2007) Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet 8:639–646
Yim SH, Kim TM, Hu HJ, Kim JH, Kim BJ, Lee JY, Han BG, Shin SH, Jung SH, Chung YJ (2010) Copy number variations in East-Asian population and their evolutionary and functional implications. Hum Mol Genet 19:1001–1008
Conrad DF, Pinto D, Redon R et al (2010) Origins and functional impact of copy number variation in the human genome. Nature 464:704–712
Egan CM, Sridhar S, Wigler M, Hall IM (2007) Recurrent DNA copy number variation in the laboratory mouse. Nat Genet 39:1384–1389
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y et al (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949
Kidd JM et al (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453(7191):56–64
Lee AS et al (2008) Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet 17:1127–1136
Perry GH et al (2006) Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci U S A 103:8006–8011
Redon R et al (2006) Global variation in copy number in the human genome. Nature 444:444–454
Glessner JT et al (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573
Bochukova EG et al (2010) Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463:666–670
Shlien A, Malkin D (2009) Copy number variations and cancer. Genome Med 1(6):62
Chen EY, Dobrinski KP, Brown KH, Clagg R, Edelman E, Ignatius MS et al (2013) Cross-species array comparative genomic hybridization identifies novel oncogenic events in zebrafish and human embryonal rhabdomyosarcoma. PLoS Genet 9(8):e1003727
Howe K, Clark M, Stemple D (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503
Lieschke GJ, Currie PD (2007) Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8:353–367
Carpio Y, Estrada M (2006) Zebrafish as a genetic model organism. Biotecnol Apl 23:4
Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X et al (2012) Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A 109(2):529–534
Bryois J, Buil A, Evans DM, Kemp JP, Montgomery SB, Conrad DF, Ho KM, Ring S, Hurles M, Deloukas P, Davey Smith G et al (2014) Cis and trans effects of human genomic variants on gene expression. PLoS Genet 10(7):e1004461
Stranger B, Forrest M et al (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315(5813):848–853
Schadt E et al (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422:297
Chesler EJ, Lu L et al (2005) Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37(3):233–242
Stranger B, Forrest M et al (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1(6):e78
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57(1):289–300
GTEx Consortium (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45(6):580–585
Stranger B, Montgomery S et al (2012) Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8(4):e1002639
Li Q, Seo J, Stranger B et al (2013) Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152(3):633–641
Pickrell J, Marioni JC et al (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Science 464(7289):768–772
Liang L, Morar N et al (2013) A cross-platform analysis of 14 177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res 23(4):716–726
Kreimer A, Pe’er I (2013) Variants in exons and in transcription factors affect gene expression in trans. Genome Biol 14(7):R71
Cheung V, Nayak R, Wang I, Elwyn S, Cousins S, Morley M, Spielman R (2010) Polymorphic cis and trans-regulation of human gene expression. PLoS Biol 8(9):e1000480
Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO (2011) Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 21(12):2004–2013
Montgomery S, Sammeth M, Gutierrez-Arcelus M et al (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Science 464(7289):773–777
Tian L, Quitadamo A, Lin F, Shi XM (2014) Methods for population-based eQTL analysis in human genetics. Tsinghua Sci Technol 19:624–634
Acknowledgments
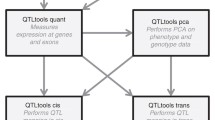
Jason Dobrinski for his assistance with Fig. 1.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Dobrinski, K.P. (2020). Tissue-Specific eQTL in Zebrafish. In: Shi, X. (eds) eQTL Analysis. Methods in Molecular Biology, vol 2082. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0026-9_17
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0026-9_17
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0025-2
Online ISBN: 978-1-0716-0026-9
eBook Packages: Springer Protocols