Abstract
Glycans play crucial roles in various biological processes such as cell proliferation, cell-cell interactions, and immune responses. Since viruses co-opt cellular biosynthetic pathways, viral glycosylation mainly depends on the host cell glycosylation machinery. Consequently, several viruses exploit the cellular glycosylation pathway to their advantage. It was shown that viral glycosylation is strongly dependent on the host system selected for virus propagation and/or protein expression. Therefore, the use of different expression systems results in various glycoforms of viral glycoproteins that may differ in functional properties. These differences clearly illustrate that the choice of the expression system can be important, as the resulting glycosylation may influence immunological properties. In this review, we will first detail protein N- and O-glycosylation pathways and the resulting glycosylation patterns; we will then discuss different aspects of viral glycosylation in pathogenesis and in vaccine development; and finally, we will elaborate on how to harness viral glycosylation in order to optimize the design of viral vaccines. To this end, we will highlight specific examples to demonstrate how glycoengineering approaches and exploitation of different expression systems could pave the way towards better self-adjuvanted glycan-based viral vaccines.
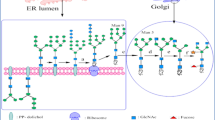
Graphical Abstract


Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- APC:
-
Antigen-presenting cell
- Asn or N:
-
Asparagine
- CHO:
-
Chinese hamster ovary
- CLR:
-
C-type lectin receptor
- DC-SIGN:
-
Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- EBOV:
-
Ebola virus
- ER:
-
Endosplasmic reticulum
- FcR:
-
Fc receptor
- FDL:
-
Fused lobes
- Fuc:
-
l-Fucose
- Gal:
-
d-Galactose
- GalNAc:
-
N-Acetyl-d-galactosamine
- Glc:
-
d-Glucose
- GlcNAc:
-
N-Acetyl-d-glucosamine
- GP:
-
Glycoprotein
- HA:
-
Hemagglutinin
- HBV:
-
Hepatitis B virus
- HCV:
-
Hepatitis C virus
- HIV-1:
-
Human immunodeficiency virus type 1
- HSV-1:
-
Herpes simplex virus type 1
- HSV-2:
-
Herpes simplex virus type 2
- JEV:
-
Japanese encephalitis virus
- LacNAc:
-
N-Acetyllactosamine (β-d-galactopyranosyl-(1→4)-2-acetamido-2-deoxy-β-d-glucopyranose)
- Man:
-
d-Mannose
- MDCK:
-
Madin-Darby canine kidney
- MDL-1:
-
Myeloid DAP12-associating lectin 1
- MMR:
-
Macrophage mannose receptor
- MPL:
-
3-O-Desacyl-4′-monophosphoryl lipid
- NA:
-
Neuraminidase
- nAb:
-
Neutralizing antibody
- Neu5Ac:
-
N-Acetylneuraminic acid
- Neu5Gc:
-
N-Glycolylneuraminic acid
- NIPV:
-
Nipah virus
- PRR:
-
Pattern recognition receptor
- RVFV:
-
Rift Valley fever phlebovirus
- Ser or S:
-
Serine
- sGP:
-
Secreted glycoprotein
- Sia:
-
Sialic acid
- SIV:
-
Simian immunodeficiency virus
- SNFG:
-
Symbol Nomenclature for Glycans
- Thr or T:
-
Threonine
- TLR:
-
Toll-like receptor
- VLP:
-
Virus-like particle
- WNV:
-
West Nile virus
- Xyl:
-
d-Xylose
References
Varki A, Gagneux P (2015) Chapter 7 – Biological functions of glycans. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 77–88
Johannssen T, Lepenies B (2017) Glycan-based cell targeting to modulate immune responses. Trends Biotechnol 35(4):334–346
Dwek RA (1996) Glycobiology: toward understanding the function of sugars. Chem Rev 96(2):683–720
Bagdonaite I, Wandall HH (2018) Global aspects of viral glycosylation. Glycobiology 28(7):443–467
Watanabe Y, Bowden TA, Wilson IA et al (2019) Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta 1863(10):1480–1497
Bagdonaite I, Vakhrushev SY, Joshi HJ et al (2018) Viral glycoproteomes: technologies for characterization and outlook for vaccine design. FEBS Lett 592(23):3898–3920
Crispin M, Doores KJ (2015) Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol 11:63–69
Stanley P, Taniguchi N, Aebi M (2015) Chapter 9 – N-glycans. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 99–111
Dicker M, Strasser R (2015) Using glyco-engineering to produce therapeutic proteins. Expert Opin Biol Ther 15(10):1501–1516
Gupta SK, Shukla P (2018) Glycosylation control technologies for recombinant therapeutic proteins. Appl Microbiol Biotechnol 102(24):10457–10468
Wang Q, Chung CY, Chough S et al (2018) Antibody glycoengineering strategies in mammalian cells. Biotechnol Bioeng 115(6):1378–1393
Buettner MJ, Shah SR, Saeui CT et al (2018) Improving immunotherapy through glycodesign. Front Immunol 9:2485
Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12(4):43R–56R
Corfield A (2017) Eukaryotic protein glycosylation: a primer for histochemists and cell biologists. Histochem Cell Biol 147(2):119–147
Zachara N, Akimoto Y, Hart GW (2015) Chapter 19 – the O-GlcNAc modification. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 239–251
Kornfeld R, Kornfeld S (1985) Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 54(1):631–664
Brockhausen I, Hull E, Hindsgaul O et al (1989) Control of glycoprotein synthesis. Detection and characterization of a novel branching enzyme from hen oviduct, UDP-N-acetylglucosamine:GlcNAc beta 1-6 (GlcNAc beta 1-2)Man alpha-R (GlcNAc to Man) beta-4-N-acetylglucosaminyltransferase VI. J Biol Chem 264(19):11211–11221
Taguchi T, Ogawa T, Inoue S et al (2000) Purification and characterization of UDP-GlcNAc: GlcNAcbeta 1-6(GlcNAcbeta 1-2)Manalpha 1-R [GlcNAc to Man]-beta 1, 4-N-acetylglucosaminyltransferase VI from hen oviduct. J Biol Chem 275(42):32598–32602
Watanabe T, Ihara H, Miyoshi E et al (2006) A specific detection of GlcNAcbeta1-6Manalpha1 branches in N-linked glycoproteins based on the specificity of N-acetylglucosaminyltransferase VI. Glycobiology 16(5):431–439
Nakano M, Mishra SK, Tokoro Y et al (2019) Bisecting GlcNAc is a general suppressor of terminal modification of N-glycan. Mol Cell Proteomics 18(10):2044–2057
Schneider M, Al-Shareffi E, Haltiwanger RS (2017) Biological functions of fucose in mammals. Glycobiology 27(7):601–618
Montero-Morales L, Steinkellner H (2018) Advanced plant-based glycan engineering. Front Bioeng Biotechnol 6(81):81
Shi X, Jarvis DL (2007) Protein N-glycosylation in the baculovirus-insect cell system. Curr Drug Targets 8(10):1116–1125
Tiemeyer M, Nakato H, Esko JD (2015) Chapter 26 – Arthropoda. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 335–349
Haltiwanger RS, Wells L, Freeze HH et al (2015) Chapter 13 – Other classes of eukaryotic glycans. In: Varki A, Cummings RD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 151–160
Varki A, Kornfeld S (2015) Chapter 1 – Historical background and overview. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 1–18
Jensen PH, Kolarich D, Packer NH (2010) Mucin-type O-glycosylation - putting the pieces together. FEBS J 277(1):81–94
Marth JD (1999) Chapter 8 – O-glycans. In: Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J (eds) Essentials of glycobiology, 1st edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Brockhausen I, Stanley P (2015) Chapter 10 – O-GalNAc glycans. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 113–123
Corfield AP, Berry M (2015) Glycan variation and evolution in the eukaryotes. Trends Biochem Sci 40(7):351–359
Stanley P, Cummings RD (2015) Chapter 14 – Structures common to different glycans. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 161–178
Fenouillet E, Gluckman JC, Bahraoui E (1990) Role of N-linked glycans of envelope glycoproteins in infectivity of human immunodeficiency virus type 1. J Virol 64(6):2841–2848
Vigerust DJ, Shepherd VL (2007) Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 15(5):211–218
Lin G, Simmons G, Pohlmann S et al (2003) Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol 77(2):1337–1346
Lozach P-Y, Amara A, Bartosch B et al (2004) C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J Biol Chem 279(31):32035–32045
Leger P, Tetard M, Youness B et al (2016) Differential use of the C-type lectins L-SIGN and DC-SIGN for phlebovirus endocytosis. Traffic 17(6):639–656
Lozach PY, Kuhbacher A, Meier R et al (2011) DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10(1):75–88
Monteiro J, Lepenies B (2017) Myeloid C-type lectin receptors in viral recognition and antiviral immunity. Viruses 9(3):59
van Liempt E, Bank CM, Mehta P et al (2006) Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett 580(26):6123–6131
Mitchell DA, Fadden AJ, Drickamer K (2001) A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. J Biol Chem 276(31):28939–28945
Curtis BM, Scharnowske S, Watson AJ (1992) Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc Natl Acad Sci U S A 89(17):8356–8360
Alvarez CP, Lasala F, Carrillo J et al (2002) C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 76(13):6841–6844
Simmons G, Reeves JD, Grogan CC et al (2003) DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305(1):115–123
Geijtenbeek TBH, Torensma R, Van Vliet SJ et al (2000) Identification of DC-SIGN, a novel dendritic cell–specific ICAM-3 receptor that supports primary immune responses. Cell 100(5):575–585
Hong PW, Flummerfelt KB, de Parseval A et al (2002) Human immunodeficiency virus envelope (gp120) binding to DC-SIGN and primary dendritic cells is carbohydrate dependent but does not involve 2G12 or cyanovirin binding sites: implications for structural analyses of gp120-DC-SIGN binding. J Virol 76(24):12855–12865
Nguyen DG, Hildreth JEK (2003) Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur J Immunol 33(2):483–493
Lai J, Bernhard OK, Turville SG et al (2009) Oligomerization of the macrophage mannose receptor enhances gp120-mediated binding of HIV-1. J Biol Chem 284(17):11027–11038
Miller JL, Dewet BJM, Martinez-Pomares L et al (2008) The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog 4(2):e17
Altgärde N, Eriksson C, Peerboom N et al (2015) Mucin-like region of herpes simplex virus type 1 attachment protein glycoprotein C (gC) modulates the virus-glycosaminoglycan interaction. J Biol Chem 290(35):21473–21485
Stone JA, Nicola AV, Baum LG et al (2016) Multiple novel functions of henipavirus O-glycans: the first O-glycan functions identified in the paramyxovirus family. PLoS Pathog 12(2):e1005445
Luo S, Hu K, He S et al (2015) Contribution of N-linked glycans on HSV-2 gB to cell–cell fusion and viral entry. Virology 483:72–82
Ito K, Qin Y, Guarnieri M et al (2010) Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol 84(24):12850–12861
Volchkov VE, Feldmann H, Volchkova VA et al (1998) Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A 95(10):5762–5767
Wang B, Wang Y, Frabutt DA et al (2017) Mechanistic understanding of N-glycosylation in Ebola virus glycoprotein maturation and function. J Biol Chem 292(14):5860–5870
Szakonyi G, Klein MG, Hannan JP et al (2006) Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol 13(11):996–1001
Sommerstein R, Flatz L, Remy MM et al (2015) Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog 11(11):e1005276
Falkowska E, Kajumo F, Garcia E et al (2007) Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol 81(15):8072–8079
Beniac DR, Booth TF (2017) Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci Rep 7:46374
Seabright GE, Doores KJ, Burton DR et al (2019) Protein and glycan mimicry in HIV vaccine design. J Mol Biol 431(12):2223–2247
Hallenberger S, Bosch V, Angliker H et al (1992) Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360(6402):358–361
Silver ZA, Antonopoulos A, Haslam SM et al (2020) Discovery of O-linked carbohydrate on HIV-1 envelope and its role in shielding against one category of broadly neutralizing antibodies. Cell Rep 30(6):1862–1869.e1864
Lasky LA, Groopman JE, Fennie CW et al (1986) Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233(4760):209–212
Lee JH, Ozorowski G, Ward AB (2016) Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351(6277):1043–1048
Stewart-Jones GB, Soto C, Lemmin T et al (2016) Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165(4):813–826
Wei X, Decker JM, Wang S et al (2003) Antibody neutralization and escape by HIV-1. Nature 422(6929):307–312
Moore PL, Gray ES, Wibmer CK et al (2012) Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18(11):1688–1692
Dacheux L, Moreau A, Ataman-Onal Y et al (2004) Evolutionary dynamics of the glycan shield of the human immunodeficiency virus envelope during natural infection and implications for exposure of the 2G12 epitope. J Virol 78(22):12625–12637
Wagh K, Kreider EF, Li Y et al (2018) Completeness of HIV-1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep 25(4):893–908.e897
McCaffrey RA, Saunders C, Hensel M et al (2004) N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol 78(7):3279–3295
Koch M, Pancera M, Kwong PD et al (2003) Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313(2):387–400
Li Y, Cleveland B, Klots I et al (2008) Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol 82(2):638–651
Back NK, Smit L, De Jong JJ et al (1994) An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199(2):431–438
Lynch RM, Wong P, Tran L et al (2015) HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol 89(8):4201–4213
Aguilar HC, Matreyek KA, Filone CM et al (2006) N-Glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80(10):4878–4889
Julithe R, Abou-Jaoude G, Sureau C (2014) Modification of the hepatitis B virus envelope protein glycosylation pattern interferes with secretion of viral particles, infectivity, and susceptibility to neutralizing antibodies. J Virol 88(16):9049–9059
Lennemann NJ, Rhein BA, Ndungo E et al (2014) Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 5(1):e00862–e00813
Sodora DL, Cohen GH, Eisenberg RJ (1989) Influence of asparagine-linked oligosaccharides on antigenicity, processing, and cell surface expression of herpes simplex virus type 1 glycoprotein D. J Virol 63(12):5184–5193
Hobman TC, Qiu ZY, Chaye H et al (1991) Analysis of rubella virus E1 glycosylation mutants expressed in COS cells. Virology 181(2):768–772
Fournillier A, Wychowski C, Boucreux D et al (2001) Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J Virol 75(24):12088–12097
Helle F, Vieyres G, Elkrief L et al (2010) Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 84(22):11905–11915
Liu M, Chen H, Luo F et al (2007) Deletion of N-glycosylation sites of hepatitis C virus envelope protein E1 enhances specific cellular and humoral immune responses. Vaccine 25(36):6572–6580
Sattentau QJ, Moore JP (1995) Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med 182(1):185–196
Sanders RW, Derking R, Cupo A et al (2013) A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9(9):e1003618
Mohan GS, Li W, Ye L et al (2012) Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog 8(12):e1003065
Moore PL, Crooks ET, Porter L et al (2006) Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol 80(5):2515–2528
Trefry JC, Wollen SE, Nasar F et al (2015) Ebola virus infections in nonhuman primates are temporally influenced by glycoprotein poly-U editing site populations in the exposure material. Viruses 7(12):6739–6754
Maruyama T, Parren PW, Sanchez A et al (1999) Recombinant human monoclonal antibodies to Ebola virus. J Infect Dis 179(s1):S235–S239
Druar C, Saini SS, Cossitt MA et al (2005) Analysis of the expressed heavy chain variable-region genes of Macaca fascicularis and isolation of monoclonal antibodies specific for the Ebola virus’ soluble glycoprotein. Immunogenetics 57(10):730–738
Cook JD, Lee JE (2013) The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog 9(5):e1003258
WHO (2019) Ten threats to global health in 2019. https://www.who.int/news-room/feature-stories/ten-threats-to-global-health-in-2019. Accessed 20 Apr 2020
Tognotti E (2010) The eradication of smallpox, a success story for modern medicine and public health: what lessons for the future? J Infect Dev Ctries 4(5):264–266
Delany I, Rappuoli R, De Gregorio E (2014) Vaccines for the 21st century. EMBO Mol Med 6(6):708–720
Vetter V, Denizer G, Friedland LR et al (2018) Understanding modern-day vaccines: what you need to know. Ann Med 50(2):110–120
Morens DM, Holmes EC, Davis AS et al (2011) Global rinderpest eradication: lessons learned and why humans should celebrate too. J Infect Dis 204(4):502–505
Hamilton K, Baron MD, Matsuo K et al (2017) Rinderpest eradication: challenges for remaining disease free and implications for future eradication efforts. Rev Sci Tech 36(2):579–588
Minor PD (2015) Live attenuated vaccines: historical successes and current challenges. Virology 479-480:379–392
Plotkin S (2014) History of vaccination. Proc Natl Acad Sci U S A 111(34):12283–12287
Hajj Hussein I, Chams N, Chams S et al (2015) Vaccines through centuries: major cornerstones of global health. Front Public Health 3:269
Barrett ADT (2017) Yellow fever live attenuated vaccine: a very successful live attenuated vaccine but still we have problems controlling the disease. Vaccine 35(44):5951–5955
Caplen H, Peters CJ, Bishop DH (1985) Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol 66(10):2271–2277
Lokugamage N, Freiberg AN, Morrill JC et al (2012) Genetic subpopulations of Rift Valley fever virus strains ZH548 and MP-12 and recombinant MP-12 strains. J Virol 86(24):13566–13575
Ikegami T, Hill TE, Smith JK et al (2015) Rift Valley fever virus MP-12 vaccine is fully attenuated by a combination of partial attenuations in the S, M, and L segments. J Virol 89(14):7262–7276
Morrill JC, Jennings GB, Caplen H et al (1987) Pathogenicity and immunogenicity of a mutagen-attenuated Rift Valley fever virus immunogen in pregnant ewes. Am J Vet Res 48(7):1042–1047
Morrill JC, Peters CJ (2003) Pathogenicity and neurovirulence of a mutagen-attenuated Rift Valley fever vaccine in rhesus monkeys. Vaccine 21(21–22):2994–3002
Morrill JC, Mebus CA, Peters CJ (1997) Safety and efficacy of a mutagen-attenuated Rift Valley fever virus vaccine in cattle. Am J Vet Res 58(10):1104–1109
Lang Y, Li Y, Jasperson D et al (2019) Identification and evaluation of antivirals for Rift Valley fever virus. Vet Microbiol 230:110–116
Ikegami T (2019) Candidate vaccines for human Rift Valley fever. Expert Opin Biol Ther 19(12):1333–1342
Thomas Jr F, Magill T (1936) Vaccination of human subjects with virus of human influenza. Proc Soc Exp Biol Med 33(4):604–606
Salk JE, Krech U, Youngner JS et al (1954) Formaldehyde treatment and safety testing of experimental poliomyelitis vaccines. Am J Public Health Nations Health 44(5):563–570
Provost PJ, Hughes JV, Miller WJ et al (1986) An inactivated hepatitis A viral vaccine of cell culture origin. J Med Virol 19(1):23–31
Kunz C (1962) Aktiv und passive Immunoprophylaxe der Fruhsommer-Meningoencephalitis (FSME). Arzneimittelforschung 28:1806
Yamashita T, Ishikawa N, Hojo F et al (1970) Japanese encephalitis purified vaccine. II. Purity of the mouse brain vaccine purified by ultracentrifugation. Biken J 13(1):25–38
Fan YC, Chiu HC, Chen LK et al (2015) Formalin inactivation of Japanese encephalitis virus vaccine alters the antigenicity and immunogenicity of a neutralization epitope in envelope protein domain III. PLoS Negl Trop Dis 9(10):e0004167
di Tommaso A, de Magistris MT, Bugnoli M et al (1994) Formaldehyde treatment of proteins can constrain presentation to T cells by limiting antigen processing. Infect Immun 62(5):1830–1834
Ibsen PH (1996) The effect of formaldehyde, hydrogen peroxide and genetic detoxification of pertussis toxin on epitope recognition by murine monoclonal antibodies. Vaccine 14(5):359–368
Widjaja I, Wicht O, Luytjes W et al (2016) Characterization of epitope-specific anti-respiratory syncytial virus (anti-RSV) antibody responses after natural infection and after vaccination with formalin-inactivated RSV. J Virol 90(13):5965–5977
Clark TG, Cassidy-Hanley D (2005) Recombinant subunit vaccines: potentials and constraints. Dev Biol (Basel) 121:153–163
Michel ML, Tiollais P (2010) Hepatitis B vaccines: protective efficacy and therapeutic potential. Pathol Biol (Paris) 58(4):288–295
Soema PC, Kompier R, Amorij J-P et al (2015) Current and next generation influenza vaccines: formulation and production strategies. Eur J Pharm Biopharm 94:251–263
Rappuoli R, Pizza M, Del Giudice G et al (2014) Vaccines, new opportunities for a new society. Proc Natl Acad Sci U S A 111(34):12288–12293
Colgrave ML, Snelling HJ, Shiell BJ et al (2012) Site occupancy and glycan compositional analysis of two soluble recombinant forms of the attachment glycoprotein of Hendra virus. Glycobiology 22(4):572–584
Orntoft TF, Vestergaard EM (1999) Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis 20(2):362–371
Rendić D, Wilson IB, Paschinger K (2008) The glycosylation capacity of insect cells. Croat Chem Acta 81(1):7–21
Vandenborre G, Smagghe G, Ghesquiere B et al (2011) Diversity in protein glycosylation among insect species. PLoS One 6(2):e16682
Walski T, De Schutter K, Van Damme EJM et al (2017) Diversity and functions of protein glycosylation in insects. Insect Biochem Mol Biol 83:21–34
Joshi HJ, Narimatsu Y, Schjoldager KT et al (2018) SnapShot: O-glycosylation pathways across kingdoms. Cell 172(3):632–632.e632
Hill BD, Zak A, Khera E et al (2018) Engineering virus-like particles for antigen and drug delivery. Curr Protein Pept Sci 19(1):112–127
Roldão A, Mellado MCM, Castilho LR et al (2010) Virus-like particles in vaccine development. Expert Rev Vaccines 9(10):1149–1176
Krugman S (1982) The newly licensed hepatitis B vaccine. Characteristics and indications for use. JAMA 247(14):2012–2015
Pasquale A, Preiss S, Silva F et al (2015) Vaccine adjuvants: from 1920 to 2015 and beyond. Vaccine 3(2):320–343
Kool M, Fierens K, Lambrecht BN (2012) Alum adjuvant: some of the tricks of the oldest adjuvant. J Med Microbiol 61(7):927–934
Didierlaurent AM, Morel S, Lockman L et al (2009) AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J Immunol 183(10):6186–6197
Morel S, Didierlaurent A, Bourguignon P et al (2011) Adjuvant system AS03 containing α-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine 29(13):2461–2473
Cohet C, van der Most R, Bauchau V et al (2019) Safety of AS03-adjuvanted influenza vaccines: a review of the evidence. Vaccine 37(23):3006–3021
Batista-Duharte A, Martínez DT, Carlos IZ (2018) Efficacy and safety of immunological adjuvants. Where is the cut-off? Biomed Pharmacother 105:616–624
Air GM (1981) Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci U S A 78(12):7639–7643
Wu Y, Wu Y, Tefsen B et al (2014) Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22(4):183–191
Sautto GA, Kirchenbaum GA, Ross TM (2018) Towards a universal influenza vaccine: different approaches for one goal. Virol J 15(1):17
Air GM (2014) Influenza virus–glycan interactions. Curr Opin Virol 7:128–133
Epstein SL, Misplon JA, Lawson CM et al (1993) Beta 2-microglobulin-deficient mice can be protected against influenza A infection by vaccination with vaccinia-influenza recombinants expressing hemagglutinin and neuraminidase. J Immunol 150(12):5484–5493
Angeletti D, Gibbs JS, Angel M et al (2017) Defining B cell immunodominance to viruses. Nat Immunol 18(4):456–463
Doyle TM, Jaentschke B, Van Domselaar G et al (2013) The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288(25):18283–18289
Webster RG, Govorkova EA (2014) Continuing challenges in influenza. Ann N Y Acad Sci 1323(1):115–139
Erbelding EJ, Post DJ, Stemmy EJ et al (2018) A universal influenza vaccine: the strategic plan for the National Institute of allergy and infectious diseases. J Infect Dis 218(3):347–354
Wei CJ, Crank MC, Shiver J et al (2020) Next-generation influenza vaccines: opportunities and challenges. Nat Rev Drug Discov 19(4):239–252
Wendel I, Matrosovich M, Klenk HD (2015) SnapShot: evolution of human influenza A viruses. Cell Host Microbe 17(3):416–416.e411
WHO (2020) Influenza (seasonal) fact sheet. https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal). Accessed 18 Apr 2020
Chang D, Zaia J (2019) Why glycosylation matters in building a better flu vaccine. Mol Cell Proteomics 18(12):2348–2358
Schwarzer J, Rapp E, Hennig R et al (2009) Glycan analysis in cell culture-based influenza vaccine production: influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine 27(32):4325–4336
Schild GC, Oxford JS, de Jong JC et al (1983) Evidence for host-cell selection of influenza virus antigenic variants. Nature 303(5919):706–709
Robertson JS, Bootman JS, Newman R et al (1987) Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A (H1N1) virus. Virology 160(1):31–37
de Vries RP, Smit CH, de Bruin E et al (2012) Glycan-dependent immunogenicity of recombinant soluble trimeric hemagglutinin. J Virol 86(21):11735–11744
An Y, Parsons LM, Jankowska E et al (2019) N-glycosylation of seasonal influenza vaccine hemagglutinins: implication for potency testing and immune processing. J Virol 93(2):e01693–e01618
Wu NC, Zost SJ, Thompson AJ et al (2017) A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog 13(10):e1006682
Granicher G, Coronel J, Pralow A et al (2019) Efficient influenza A virus production in high cell density using the novel porcine suspension cell line PBG.PK2.1. Vaccine 37(47):7019–7028
Altman MO, Angel M, Kosik I et al (2019) Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio 10(2):e00204–e00219
Li D, von Schaewen M, Wang X et al (2016) Altered glycosylation patterns increase immunogenicity of a subunit hepatitis C virus vaccine, inducing neutralizing antibodies which confer protection in mice. J Virol 90(23):10486–10498
Go EP, Ding H, Zhang S et al (2017) Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven Env trimers. J Virol 91(9):e02428–e02416
Hutter J, Rodig JV, Hoper D et al (2013) Toward animal cell culture-based influenza vaccine design: viral hemagglutinin N-glycosylation markedly impacts immunogenicity. J Immunol 190(1):220–230
Liu WC, Lin YL, Spearman M et al (2016) Influenza virus hemagglutinin glycoproteins with different N-glycan patterns activate dendritic cells in vitro. J Virol 90(13):6085–6096
Urbanowicz RA, Wang R, Schiel JE et al (2019) Antigenicity and immunogenicity of differentially glycosylated hepatitis C virus E2 envelope proteins expressed in mammalian and insect cells. J Virol 93(7):e01403–e01418
Ronda C, Pedersen LE, Hansen HG et al (2014) Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng 111(8):1604–1616
Toth AM, Kuo C-W, Khoo K-H et al (2014) A new insect cell glycoengineering approach provides baculovirus-inducible glycogene expression and increases human-type glycosylation efficiency. J Biotechnol 182-183:19–29
Heffner KM, Wang Q, Hizal DB et al (2018) Glycoengineering of mammalian expression systems on a cellular level. In: Advances in biochemical engineering/biotechnology. Springer, Berlin. https://doi.org/10.1007/1010_2017_1057
Mabashi-Asazuma H, Jarvis DL (2017) CRISPR-Cas9 vectors for genome editing and host engineering in the baculovirus-insect cell system. Proc Natl Acad Sci U S A 114(34):9068–9073
Narimatsu Y, Joshi HJ, Nason R et al (2019) An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol Cell 75(2):394–407.e395
Yang Z, Wang S, Halim A et al (2015) Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotechnol 33:842
Lepenies B, Seeberger PH (2014) Simply better glycoproteins. Nat Biotechnol 32(5):443–445
Traving C, Schauer R (1998) Structure, function and metabolism of sialic acids. Cell Mol Life Sci 54(12):1330–1349
Varki A, Gagneux P (2012) Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci 1253(1):16–36
Dhar C, Sasmal A, Varki A (2019) From “serum sickness” to “xenosialitis”: past, present, and future significance of the non-human sialic acid Neu5Gc. Front Immunol 10:807
Altman MO, Gagneux P (2019) Absence of Neu5Gc and presence of anti-Neu5Gc antibodies in humans-an evolutionary perspective. Front Immunol 10:789
Ghaderi D, Taylor RE, Padler-Karavani V et al (2010) Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol 28(8):863–867
Benatuil L, Kaye J, Rich RF et al (2005) The influence of natural antibody specificity on antigen immunogenicity. Eur J Immunol 35(9):2638–2647
Abdel-Motal UM, Wigglesworth K, Galili U (2009) Mechanism for increased immunogenicity of vaccines that form in vivo immune complexes with the natural anti-gal antibody. Vaccine 27(23):3072–3082
Bakema JE, Tuk CW, van Vliet SJ et al (2015) Antibody-opsonized bacteria evoke an inflammatory dendritic cell phenotype and polyfunctional Th cells by cross-talk between TLRs and FcRs. J Immunol 194(4):1856–1866
Huai G, Qi P, Yang H et al (2016) Characteristics of α-gal epitope, anti-gal antibody, α1,3 galactosyltransferase and its clinical exploitation (review). Int J Mol Med 37(1):11–20
Macher BA, Galili U (2008) The Galα1,3Galβ1,4GlcNAc-R (α-gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim Biophys Acta 1780(2):75–88
Abdel-Motal UM, Guay HM, Wigglesworth K et al (2007) Immunogenicity of influenza virus vaccine is increased by anti-gal-mediated targeting to antigen-presenting cells. J Virol 81(17):9131–9141
Abdel-Motal U, Wang S, Lu S et al (2006) Increased immunogenicity of human immunodeficiency virus gp120 engineered to express Galalpha1-3Galbeta1-4GlcNAc-R epitopes. J Virol 80(14):6943–6951
Abdel-Motal UM, Wang S, Awad A et al (2010) Increased immunogenicity of HIV-1 p24 and gp120 following immunization with gp120/p24 fusion protein vaccine expressing alpha-gal epitopes. Vaccine 28(7):1758–1765
Henion TR, Gerhard W, Anaraki F et al (1997) Synthesis of alpha-gal epitopes on influenza virus vaccines, by recombinant alpha-1,3-galactosyltransferase, enables the formation of immune complexes with the natural anti-gal antibody. Vaccine 15(11):1174–1182
Galili U, Repik PM, Anaraki F et al (1996) Enhancement of antigen presentation of influenza virus hemagglutinin by the natural human anti-gal antibody. Vaccine 14(4):321–328
Steinke JW, Platts-Mills TA, Commins SP (2015) The alpha-gal story: lessons learned from connecting the dots. J Allergy Clin Immunol 135(3):589–596
Chinuki Y, Morita E (2019) Alpha-gal-containing biologics and anaphylaxis. Allergol Int 68(3):296–300
Román-Carrasco P, Lieder B, Somoza V et al (2019) Only α-gal bound to lipids, but not to proteins, is transported across enterocytes as an IgE-reactive molecule that can induce effector cell activation. Allergy 74(10):1956–1968
Lepenies B, Lee J, Sonkaria S (2013) Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv Drug Del Rev 65(9):1271–1281
Brzezicka K, Vogel U, Serna S et al (2016) Influence of core beta-1,2-xylosylation on glycoprotein recognition by murine C-type lectin receptors and its impact on dendritic cell targeting. ACS Chem Biol 11(8):2347–2356
Johannssen T, Lepenies B (2015) Identification and characterization of carbohydrate-based adjuvants. Methods Mol Biol 1331:173–187
Maglinao M, Eriksson M, Schlegel MK et al (2014) A platform to screen for C-type lectin receptor-binding carbohydrates and their potential for cell-specific targeting and immune modulation. J Control Release 175:36–42
Mayer S, Raulf M-K, Lepenies B (2017) C-type lectins: their network and roles in pathogen recognition and immunity. Histochem Cell Biol 147(2):223–237
Goyal S, Castrillon-Betancur JC, Klaile E et al (2018) The interaction of human pathogenic fungi with C-type lectin receptors. Front Immunol 9:1261
van Kooyk Y, Unger WWJ, Fehres CM et al (2013) Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol Immunol 55(2):143–145
Hu J, Wei P, Seeberger PH et al (2018) Mannose-functionalized nanoscaffolds for targeted delivery in biomedical applications. Chem Asian J 13(22):3448–3459
Gemmill TR, Trimble RB (1999) Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta 1426(2):227–237
Kottom TJ, Hebrink DM, Monteiro JT et al (2019) Myeloid C-type lectin receptors that recognize fungal mannans interact with Pneumocystis organisms and major surface glycoprotein. J Med Microbiol 68(11):1649–1654
Angrand G, Quillévéré A, Loaëc N et al (2019) Sneaking out for happy hour: yeast-based approaches to explore and modulate immune response and immune evasion. Genes 10(9):667
Vetvicka V, Vannucci L, Sima P (2020) Beta-glucan as a new tool in vaccine development. Scand J Immunol 91(2):e12833
Stanley P, Chen W (2003) Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology 13(1):43–50
Byrne G, O’Rourke SM, Alexander DL et al (2018) CRISPR/Cas9 gene editing for the creation of an MGAT1-deficient CHO cell line to control HIV-1 vaccine glycosylation. PLoS Biol 16(8):e2005817
Cox MMJ, Hollister JR (2009) FluBlok, a next generation influenza vaccine manufactured in insect cells. Biologicals 37(3):182–189
Wilson IBH (2002) Glycosylation of proteins in plants and invertebrates. Curr Opin Struct Biol 12(5):569–577
Altmann F (2007) The role of protein glycosylation in allergy. Int Arch Allergy Immunol 142(2):99–115
Gaunitz S, Jin C, Nilsson A et al (2013) Mucin-type proteins produced in the Trichoplusia ni and Spodoptera frugiperda insect cell lines carry novel O-glycans with phosphocholine and sulfate substitutions. Glycobiology 23(7):778–796
Kurz S, Aoki K, Jin C et al (2015) Targeted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J Proteome 126:172–188
Wilson IBH, Cummings RD, Aebi M (2015) Chapter 25 – Nematoda. In: Varki A, Cummings RD, Esko JD et al (eds) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 321–333
Martini F, Eckmair B, Stefanic S et al (2019) Highly modified and immunoactive N-glycans of the canine heartworm. Nat Commun 10(1):75
Shim BS, Hong KJ, Maharjan PM et al (2019) Plant factory: new resource for the productivity and diversity of human and veterinary vaccines. Clin Exp Vaccine Res 8(2):136–139
Takeyama N, Kiyono H, Yuki Y (2015) Plant-based vaccines for animals and humans: recent advances in technology and clinical trials. Ther Adv Vaccine 3(5–6):139–154
D’Aoust M-A, Couture MMJ, Charland N et al (2010) The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant Biotechnol J 8(5):607–619
Landry N, Ward BJ, Trépanier S et al (2010) Preclinical and clinical development of plant-made virus-like particle vaccine against avian H5N1 influenza. PLoS One 5(12):e15559
Landry N, Pillet S, Favre D et al (2014) Influenza virus-like particle vaccines made in Nicotiana benthamiana elicit durable, poly-functional and cross-reactive T cell responses to influenza HA antigens. Clin Immunol 154(2):164–177
Le Mauff F, Mercier G, Chan P et al (2015) Biochemical composition of haemagglutinin-based influenza virus-like particle vaccine produced by transient expression in tobacco plants. Plant Biotechnol J 13(5):717–725
Margolin E, Chapman R, Williamson A-L et al (2018) Production of complex viral glycoproteins in plants as vaccine immunogens. Plant Biotechnol J 16(9):1531–1545
Ward BJ, Landry N, Trépanier S et al (2014) Human antibody response to N-glycans present on plant-made influenza virus-like particle (VLP) vaccines. Vaccine 32(46):6098–6106
Shaaltiel Y, Tekoah Y (2016) Plant specific N-glycans do not have proven adverse effects in humans. Nat Biotechnol 34(7):706–708
Rup B, Alon S, Amit-Cohen B-C et al (2017) Immunogenicity of glycans on biotherapeutic drugs produced in plant expression systems—the taliglucerase alfa story. PLoS One 12(10):e0186211
Smith DF, Cummings RD, Song X (2019) History and future of shotgun glycomics. Biochem Soc Trans 47(1):1–11
Tamburrini A, Colombo C, Bernardi A (2019) Design and synthesis of glycomimetics: recent advances. Med Res Rev 40(2):495–531
Guan WJ, Ni ZY, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. https://doi.org/10.1056/NEJMoa2002032
Wu F, Zhao S, Yu B et al (2020) A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269
Varki A, Cummings RD, Aebi M et al (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25(12):1323–1324
Hastie KM, Zandonatti MA, Kleinfelter LM et al (2017) Structural basis for antibody-mediated neutralization of Lassa virus. Science 356(6341):923–928
Watanabe Y, Raghwani J, Allen JD et al (2018) Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc Natl Acad Sci U S A 115(28):7320–7325
Zhao Y, Ren J, Harlos K et al (2016) Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 535(7610):169–172
Lee PS, Ohshima N, Stanfield RL et al (2014) Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5(1):3614
An Y, McCullers JA, Alymova I et al (2015) Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J Proteome Res 14(9):3957–3969
Struwe WB, Chertova E, Allen JD et al (2018) Site-specific glycosylation of virion-derived HIV-1 Env is mimicked by a soluble trimeric immunogen. Cell Rep 24(8):1958–1966.e1955
Kwon YD, Pancera M, Acharya P et al (2015) Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22(7):522–531
Walls AC, Tortorici MA, Frenz B et al (2016) Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23(10):899–905
Xu K, Chan YP, Bradel-Tretheway B et al (2015) Crystal structure of the pre-fusion Nipah virus fusion glycoprotein reveals a novel hexamer-of-trimers assembly. PLoS Pathog 11(12):e1005322
Acknowledgments
G. Goyette-Desjardins is a recipient of a postdoctoral research fellowship from the “Fonds de recherche du Québec - Nature et technologies” (FRQNT, Canada). K. Schön is funded by the “Deutsche Forschungsgemeinschaft” (DFG, Germany; #398066876/GRK 2485/1).
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Schön, K., Lepenies, B., Goyette-Desjardins, G. (2020). Impact of Protein Glycosylation on the Design of Viral Vaccines. In: Rapp, E., Reichl, U. (eds) Advances in Glycobiotechnology. Advances in Biochemical Engineering/Biotechnology, vol 175. Springer, Cham. https://doi.org/10.1007/10_2020_132
Download citation
DOI: https://doi.org/10.1007/10_2020_132
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-69589-7
Online ISBN: 978-3-030-69590-3
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)