Abstract
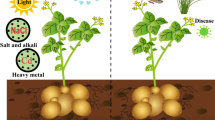
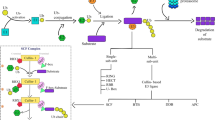
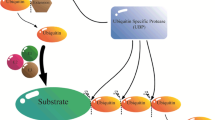
Post-translational modifications namely ubiquitination, phosphorylation, methylation and acetylation play distinct roles in regulating the growth and development of plants. Among these, the ubiquitination regulates the abundance, activities, subcellular compartmentalization and trafficking of regulatory proteins involved in diverse developmental as well as stress-responsive processes. The ubiquitin–proteasome system (UPS) involves five essential components namely ubiquitin, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin ligase (E3) and the intact 26S proteasome. The E3 ubiquitin ligase is the major component of UPS that recognizes and tethers poly-ubiquitins on the target proteins. Owing to its specificity of substrate recognition, the E3 ubiquitin ligase contributes not only to the proteome plasticity of the cell but also regulates the plant’s response to environmental cues. In this context, the review summarizes the components involved in UPS and elaborates the role of E3 ubiquitin ligase in biotic and abiotic stress responses.

Similar content being viewed by others
References
Adams EHS, Spoel SH. The ubiquitin–proteasome system as a transcriptional regulator of plant immunity. J Exp Bot. 2018;69:4529–37.
Adler G, Konrad Z, Zamir L, Mishra AK, Raveh D, Bar-Zvi D. The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 2017;17:8.
Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, et al. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol. 2005;59:469–84.
Aragon W, Reina-Pinto JJ, Serrano M. The intimate talk between plants and microorganisms at the leaf surface. J Exp Bot. 2017;68:5339–50.
Aravind L, Koonin EV. The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol. 2000;10:R132–4.
Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6:354–8.
Blanc C, Coluccia F, L’Haridon F, Torres M, Ortiz-Berrocal M, Stahl E, et al. The cuticle mutant eca2 modifies plant defense responses to biotrophic and necrotrophic pathogens and herbivory insects. Mol Plant Microbe Interact. 2018;31:344–55.
Bonas U, Lahaye T. Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr Opin Microbiol. 2002;5:44–50.
Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–54.
Callis J, Carpenter T, Sun CW, Vierstra RD. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 1995;139:921–39.
Chinnusamy V. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–54.
Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–914.
Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–89.
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA. 2006;103:8281–6.
Downes B, Vierstra RD. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans. 2005;33:393–9.
Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin–protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003;35:729–42.
Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822.
Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–92.
Genschik P, Parmentier Y, Durr A, Marbach J, Criqui MC, Jamet E, et al. Ubiquitin genes are differentially regulated in protoplast-derived cultures of Nicotiana sylvestris and in response to various stresses. Plant Mol Biol. 1992;20:897–910.
Ghannam A, Jacques A, de Ruffray P, Kauffmann S. NtRING1, putative RING-finger E3 ligase protein, is a positive regulator of the early stages of elicitin-induced HR in tobacco. Plant Cell Rep. 2016;35:415–28.
Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–9.
Hatfield PM, Gosink MM, Carpenter TB, Vierstra RD. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 1997;11:213–26.
He F, Wang HL, Li HG, Su Y, Li S, Yang Y, et al. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol J. 2018;16:1514–28.
Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–34.
Heise A, Lippok B, Kirsch C, Hahlbrock K. Two immediate-early pathogen-responsive members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proc Natl Acad Sci USA. 2002;99:9049–54.
Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin–protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–86.
Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10.
Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K. A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J. 2001;26:217–27.
Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005;139(4):1597–611.
Kurepa J, Smalle JA. Structure, function and regulation of plant proteasomes. Biochimie. 2008;90:324–35.
Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell. 2009;21:622–41.
Lee JC, Peter ME. Regulation of apoptosis by ubiquitination. Immunol Rev. 2003;193:39–47.
Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487.
Lim SD, Cho HY, Park YC, Ham DJ, Lee JK, Jang CS. The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J Exp Bot. 2013;64:2899–914.
Lim SD, Hwang JG, Jung CG, Hwang SG, Moon JC, Jang CS. Comprehensive analysis of the rice RING E3 ligase family reveals their functional diversity in response to abiotic stress. DNA Res. 2013;20:299–314.
Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–7.
Luo H, Laluk K, Lai Z, Veronese P, Song F, Mengiste T. The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 2010;154:1766–82.
Mandal A, Mishra AK, Dulani P, Muthamilarasan M, Shweta S, Prasad M. Identification, characterization, expression profiling, and virus-induced gene silencing of armadillo repeat-containing proteins in tomato suggest their involvement in tomato leaf curl New Delhi virus resistance. Funct Integr Genomics. 2018;18:101–11.
Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, et al. Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nature Commun. 2013;4:1476.
Marino D, Peeters N, Rivas S. Ubiquitination during plant immune signaling. Plant Physiol. 2012;160:15–27.
Martin T. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon: nitrogen availability. Plant Physiol. 2002;128:472–81.
Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, et al. The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics. 2006;7:509–22.
Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15:2551–65.
Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. J Exp Bot. 2007;58:2297–306.
Molnar G, Bancos S, Nagy F, Szekeres M. Characterisation of BRH1, a brassinosteroid-responsive RING-H2 gene from Arabidopsis thaliana. Planta. 2002;215:127–33.
Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134:59–66.
Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–5.
Muthamilarasan M, Prasad M. Plant innate immunity: an updated insight into defense mechanism. J Biosci. 2013;38:433–49.
Ning Y, Jantasuriyarat C, Zhao Q, Zhang H, Chen S, Liu J, et al. The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 2011;157:242–55.
Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–66.
Park J-A, Cho SK, Kim JE, Chung HS, Hong J-P, Hwang B, et al. Isolation of cDNAs differentially expressed in response to drought stress and characterization of the Ca-LEAL1 gene encoding a new family of atypical LEA-like protein homologue in hot pepper (Capsicum annuum L. cv. Pukang). Plant Sci. 2003;165:471–81.
Peng M, Hannam C, Gu H, Bi Y-M, Rothstein SJ. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007;50:320–37.
Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6.
Pokhilko A, Ramos JA, Holtan H, Maszle DR, Khanna R, Millar AJ. Ubiquitin ligase switch in plant photomorphogenesis: a hypothesis. J Theor Biol. 2011;270:31–41.
Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–707.
Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20:752–67.
Rowland O, Ludwig AA, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, et al. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell. 2005;17:295–310.
Sahu PP, Sharma N, Puranik S, Chakraborty S, Prasad M. Tomato 26S Proteasome subunit RPT4a regulates ToLCNDV transcription and activates hypersensitive response in tomato. Sci Rep. 2016;6:270–8.
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA. 2006;103:18822–7.
Salinas-Mondragon RE, Garciduenas-Pina C, Guzman P. Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol Biol. 1999;40:579–90.
Sato T, Maekawa S, Yasuda S, Sonoda Y, Katoh E, Ichikawa T, et al. CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 2009;60:852–64.
Serrano M, Guzman P. Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics. 2004;167:919–29.
Shen Q, Hu T, Bao M, Cao L, Zhang H, Song F, et al. Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded betaC1. Mol Plant. 2016;9:911–25.
Shu K, Yang W. E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017;58:1461–76.
Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–90.
Sonoda Y, Sako K, Maki Y, Yamazaki N, Yamamoto H, Ikeda A, et al. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. Plant J. 2009;60:68–78.
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30.
Sun CW, Callis J. Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 1997;11:1017–27.
Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, et al. EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J. 2002;30:447–55.
Takizawa M, Goto A, Watanabe Y. The tobacco ubiquitin-activating enzymes NtE1A and NtE1B are induced by tobacco mosaic virus, wounding and stress hormones. Mol Cells. 2005;19:228–31.
Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, et al. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 2005;43:437–48.
Thordal-Christensen H. Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol. 2003;6:351–7.
Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102.
Trujillo M, Shirasu K. Ubiquitination in plant immunity. Curr Opin Plant Biol. 2010;13:402–8.
Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302.
Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97.
Wiborg J, O’Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin–protein ligases. Biochem J. 2008;413:447–57.
Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–9.
Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–21.
Yu Y, Xu W, Wang J, Wang L, Yao W, Yang Y, et al. The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 2013;200:834–46.
Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, et al. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–808.
Zhang C, Song L, Choudhary MK, Zhou B, Sun G, Broderick K, Gieslet L, Zeng L. Genome-wide analysis of genes encoding core components of the ubiquitin system in soybean (Glycine max) reveals a potential role for ubiquitination in host immunity against soybean cyst nematode. BMC Plant Biol. 2018;18:149.
Acknowledgments
Authors’ work in this area is financially supported by the Department of Biotechnology, Ministry of Science and Technology, Govt. of India (Project No.: BT/PR8357/PBD/16/1033/2013). AM acknowledges the NPDF award from DST-SERB, Govt. of India. MM acknowledges the DST INSPIRE Faculty Award from DST, Govt. of India. The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to the e-resources.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is dedicated to the memory of Profs AK Sharma and Archana Sharma.
Rights and permissions
About this article
Cite this article
Mandal, A., Sharma, N., Muthamilarasan, M. et al. Ubiquitination: a tool for plant adaptation to changing environments. Nucleus 61, 253–260 (2018). https://doi.org/10.1007/s13237-018-0255-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-018-0255-6