Abstract
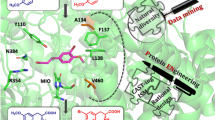
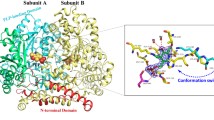
Phenylalanine ammonia lyase from Anabaena variabilis (Av-PAL) is a candidate for the treatment of phenylketonuria (PKU). However, Av-PAL shows its optimal pH at 8.5 and maintains only 70% of its highest activity when pH decreases to 7.3–7.4 (the condition of human plasma). The objective of the study was to shift its optimal pH by mutating surface amino acid residues which interact with the general base Tyr78. Based on the crystal structure and the online program GETAREA, we selected five sites: Asn69, Glu72, Glu75, Asn89, and Val90. Removing negative charges or introducing positive charges near the general base Tyr78 by mutation, the pH optima were successfully shifted to more acidic range. Especially, the pH optima of E75A, E75L, and E75Q were shifted to 7.5 with 35, 30, and 24% higher specific activities than that of the wild, respectively. Half-lives of E75L and E75Q at 70 °C prolonged to 190 and 180 min from 130 min of the wild, respectively. In addition, the higher resistance to a low pH of 3.5 and protease made E75L a candidate for oral medicine of PKU. This work would improve the therapeutic prospect of Av-PAL and provide guidance in modulating optimal pH of enzymes.






Similar content being viewed by others
Reference
Giberti, S., Bertea, C. M., Narayana, R., Maffei, M. E., & Forlani, G. (2012). Two phenylalanine ammonia lyase isoforms are involved in the elicitor-induced response of rice to the fungal pathogen Magnaporthe oryzae. Journal of Plant Physiology, 169, 249–254.
Hou, X., Shao, F., Ma, Y., & Lu, S. (2013). The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Molecular Biology Reports, 40, 4301–4310.
Jin, Q., Yao, Y., Cai, Y., & Lin, Y. (2013). Molecular cloning and sequence analysis of a phenylalanine ammonia-lyase gene from Dendrobium. PloS One, 8, e62352.
Shang, Q. M., Li, L., & Dong, C. J. (2012). Multiple tandem duplication of the phenylalanine ammonia lyase genes in Cucumis. sativus L. Planta, 236, 1093–1105.
Wang, X. H., Gong, M., Tang, L., Zheng, S., Lou, J. D., Ou, L., Gomes-Laranjo, J., & Zhang, C. (2013). Cloning, bioinformatics and the enzyme activity analyses of a phenylalanine ammonia-lyase gene involved in dragon’s blood biosynthesis in Dracaena cambodiana. Molecular Biology Reports, 40, 97–107.
Gilbert, H. J., Clarke, I. N., Gibson, R. K., Stephenson, J. R., & Tully, M. (1985). Molecular cloning of the phenylalanine ammonia lyase gene from Rhodosporidium toruloides in Escherichia coli K-12. Journal of Bacteriology, 161, 314–320.
Zhu, L. B., Cui, W. J., Fang, Y. Q., Liu, Y., Gao, X. X., & Zhou, Z. M. (2013). Cloning, expression and characterization of phenylalanine ammonia-lyase from Rhodotorula glutinis. Biotechnology Letters, 35, 751–756.
Kim, M., Yoon, H., You, Y. H., Kim, Y. E., Woo, J. R., Seo, Y., Lee, G. M., Kim, Y. J., Kong, W. S., & Kim, J. G. (2013). Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. Journal of Microbiology and Biotechnology, 23, 1347–1356.
Vaslet, C. A., Strausberg, R. L., Sykes, A., Levy, A., & Filpula, D. (1988). cDNA and genomic cloning of yeast phenylalanine ammonia-lyase genes reveal genomic intron deletions. Nucleic Acids Research, 16, 11382.
Moffitt, M. C., Louie, G. V., Bowman, M. E., Pence, J., Noel, J. P., & Moore, B. S. (2007). Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry, 46, 1004–1012.
Kovács, K., Bánóczi, G., Varga, A., Szabó, I., Holczinger, A., Hornyánszky, G., Zagyva, I., Paizs, C., Vértessy, B. G., & Poppe, L. (2014). Expression and properties of the highly alkalophilic phenylalanine ammonia-lyase of thermophilic Rubrobacter xylanophilus. PloS One, 9, e85943–e85943.
Williams, J. S., Thomas, M., & Clarke, D. J. (2005). The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibiotic in Photorhabdus luminescens TT01. Microbiology, 151, 2543–2550.
Fowler, Z. L., & Koffas, M. A. G. (2009). Biosynthesis and biotechnological production of flavanones: current state and perspectives. App. Microbiol. Biot., 83, 799–808.
Horinouchi, S. (2009). Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Current Opinion in Chemical Biology, 13, 197–204.
Kong, J. Q. (2015). Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Advances, 5, 62587–62603.
Babich, O. O., Pokrovsky, V. S., Anisimova, N. Y., Sokolov, N. N., & Prosekov, A. Y. (2013). Recombinant L-phenylalanine ammonia lyase from Rhodosporidium toruloides as a potential anticancer agent. Biotechnol. Appl. Bioc., 60, 316–322.
Shen, R. S., Fritz, R. R., & Abell, C. W. (1977). Clearance of phenylalanine ammonia-lyase from normal and tumor-bearing mice. Cancer Research, 37, 1051–1056.
Jaliani, H. Z., Farajnia, S., Mohammadi, S. A., Barzegar, A., & Talebi, S. (2013). Engineering and kinetic stabilization of the therapeutic enzyme Anabeana variabilis phenylalanine ammonia lyase. Applied Biochemistry and Biotechnology, 171, 1805–1818.
Longo, N., Harding, C. O., Burton, B. K., Grange, D. K., Vockley, J., Wasserstein, M., Dorenbaum, A., Neuenburg, J. K., & Musson, D. G. (2014). Single-dose, subcutaneous recombinant phenylalanine ammonia lyase conjugated with polyethylene glycol in adult patients with phenylketonuria: an open-label, multicentre, phase 1 dose-escalation trial. Lancet, 384, 37–44.
Sarkissian, C. N., Kang, T. S., Gámez, A., Scriver, C. R., & Stevens, R. C. (2011). Evaluation of orally administered PEGylated phenylalanine ammonia lyase in mice for the treatment of phenylketonuria. Molecular Genetics and Metabolism, 104, 249–254.
Manikandan, K., Bhardwaj, A., Gupta, N., Lokanath, N. K., Ghosh, A., Reddy, V. S., & Ramakumar, S. (2006). Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions. Protein Science, 15, 1951–1960.
Cockburn, D. W., & Clarke, A. J. (2011). Modulating the pH-activity profile of cellulase A from Cellulomonas fimi by replacement of surface residues. Protein Engineering, Design & Selection, 24, 429–437.
Tomschy, A., Brugger, R., Lehmann, M., Svendsen, A., Vogel, K., Kostrewa, D., Lassen, S. F., Burger, D., Kronenberger, A., van Loon, A. P. G. M., Pasamontes, L., & Wyss, M. (2002). Engineering of phytase for improved activity at low pH. Appl. Environ. Microb., 68, 1907–1913.
Kim, T., Mullaney, E. J., Porres, J. M., Roneker, K. R., Crowe, S., Rice, S., Ko, T., Ullah, A. H. J., Daly, C. B., Welch, R., & Lei, X. G. (2006). Shifting the pH profile of Aspergillus niger PhyA phytase to match the stomach pH enhances its effectiveness as an animal feed additive. Appl. Environ. Microb., 72, 4397–4403.
Hirata, A., Adachi, M., Utsumi, S., & Mikami, B. (2004). Engineering of the pH optimum of Bacillus cereus β-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type. Biochemistry, 43, 12523–12531.
Cockburn, D. W., Vandenende, C., & Clarke, A. J. (2010). Modulating the pH-activity profile of cellulase by substitution: replacing the general base catalyst aspartate with cysteinesulfinate in cellulase A from Cellulomonas fimi. Biochemistry, 49, 2042–2050.
Siddiqui, K. S., Lovinyanderton, T., Rangarajan, M., & Hartley, B. S. (1993). Arthrobacter D-xylose isomerase: chemical modification of carboxy groups and protein engineering of pH optimum. The Biochemical Journal, 296, 685–691.
Cooke, H. A., Christianson, C. V., & Bruner, S. D. (2009). Structure and chemistry of 4-methylideneimidazole-5-one containing enzymes. Current Opinion in Chemical Biology, 13, 460–468.
Fraczkiewicz, R., & Braun, W. (1998). Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. Journal of Computational Chemistry, 19, 319–333.
Fiser, A., & Šali, A. (2003). Modeller: generation and refinement of homology-based protein structure models. Method. Enzymol., 374, 461–491.
Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S., & Karplus, M. (1983). CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry, 4, 187–217.
Calabrese, J. C., Jordan, D. B., Boodhoo, A., Sariaslani, S., & Vannelli, T. (2004). Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry, 43, 11403–11416.
Pilbák, S., Tomin, A., Rétey, J., & Poppe, L. (2006). The essential tyrosine-containing loop conformation and the role of the C-terminal multi-helix region in eukaryotic phenylalanine ammonia-lyases. The FEBS Journal, 273, 1004–1019.
Ostanin, K., Harms, E. H., Stevis, P. E., Kuciel, R., Zhou, M. M., & Van Etten, R. L. (1992). Overexpression, site-directed mutagenesis, and mechanism of Escherichia coli acid phosphatase. J. Bio. Chem., 267, 22830–22836.
Kang, T. S., Wang, L., Sarkissian, C. N., Gamez, A., Scriver, C. R., & Stevens, R. C. (2010). Converting an injectable protein therapeutic into an oral form: phenylalanine ammonia lyase for phenylketonuria. Molecular Genetics and Metabolism, 99, 4–9.
Jr, K. J., Nyberg, K., Sali, D., & Fersht, A. R. (2011). Contribution of hydrophobic interactions to protein stability. Journal of Molecular Biology, 408, 514–528.
Acknowledgements
This work was mainly supported by Key Laboratory of Industrial Biotechnology, Ministry of Education, China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding Sources
This work was partly funded by the National Natural Science Foundation of China (31300087, 31400058, 31671797, and 21506172), the Natural Science Foundation of Jiangsu Province of China (BK20130131, BK20130139, and BK20140151), the National High Technology Research and Development Program of China (863 Program, 2014AA021304), the High Foreign Experts Project (GDW20123200114), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (111-2-06), the Jiangsu Province “Collaborative Innovation Center for Advanced Industrial Fermentation” Industry Development Program and the Fundamental Research Funds for the Central Universities (JUSRP51411B, JUSRP51504, JUSRP51611A), and the Natural Science Foundation of Anhui Province University of China (KJ2016A801).
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
(DOCX 164 kb)
Rights and permissions
About this article
Cite this article
Zhang, F., Huang, N., Zhou, L. et al. Modulating the pH Activity Profiles of Phenylalanine Ammonia Lyase from Anabaena variabilis by Modification of Center-Near Surface Residues. Appl Biochem Biotechnol 183, 699–711 (2017). https://doi.org/10.1007/s12010-017-2458-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2458-8