Abstract
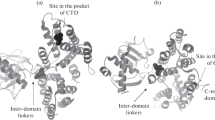
In Lactococcus lactis, the global transcriptional regulatory factor CodY can interact with the promoter DNA to regulate the growth, metabolism, environmental adaptation and other biological activities of the strains. In order to study the mechanism of interaction between CodY and its target DNA, molecular docking and molecular dynamics simulations were used to explore the binding process at molecular level. Through the calculations of the free energy of binding, hydrogen bonding and energy decomposition, nine key residues of CodY were identified, corresponding to SER184, SER186, SER208, THR217, ARG218, SER219, ASN223, LYS242 and GLY243, among which SER186, ARG218 and LYS242 play a vital role in DNA binding. Our research results provide important theoretical guidance for using wet-lab methods to study and optimize the metabolic network regulated by CodY.

Similar content being viewed by others
References
Zhang J, Zhong J. The journey of nisin development in China, a natural-green food preservative. Protein & Cell, 2015, 6(10): 709–711
Song A, In L, Lim S, Rahim R A. A review on Lactococcus lactis: From food to factory. Microbial Cell Factories, 2017, 16(1): 55
Papadimitriou K, Alegria A, Bron P A, Angelis M, Gobbetti M, Kleerebezem M. Stress physiology of lactic acid bacteria. Microbiology and Molecular Biology Reviews, 2016, 80(3): 837–890
McMahon D J, Oberg C J, Drake M A, Farkye N, Moyes L V, Arnold M R, Ganesan B, Steele J, Broadbent J R. Effect of sodium, potassium, magnesium, and calcium salt cations on pH, proteolysis, organic acids, and microbial populations during storage of full-fat cheddar cheese. Journal of Dairy Science, 2014, 97(8): 4780–4798
Laroute V, Yasaro C, Narin W, Mazzoli R, Pessione E, Loubiere P. GABA production in Lactococcus lactis is enhanced by arginine and co-addition of malate. Frontiers in Microbiology, 2016, 7: 1050
Delcour J, Ferain T, Deghorain M, Palumbo E, Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek, 1999, 76(1/4): 159–184
Hartke A, Bouche S, Giard J C, Benachour A, Boutibonnes P, Auffray Y. The lactic acid stress response of Lactococcus lactis subsp. lactis. Current Microbiology, 1996, 33(3): 194–199
Belitsky B R, Sonenshein A L. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(17): 7026–7031
Yuan T, Guo Y K, Dong J K, Li T Y, Zhou T, Sun K W, Zhang M, Wu Q Y, Xie Z, Cai Y Z, et al. Construction, characterization and application of a genome-wide promoter library in Saccharomyces cerevisiae. Frontiers of Chemical Science and Engineering, 2017, 11 (1): 107–116
Levdikov V M, Blagova E, Young V L, Belitsky B R, Lebedev A, Sonenshein A L, Wilkinson A J. Structure of the branched-chain amino acid and GTP-sensing global regulator, CodY, from Bacillus subtilis. Journal of Biological Chemistry, 2017, 292(7): 2714–2728
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Klefer F, Cassarino T G, Bertonl M, Bordoli L, et al. Swissmodel: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 2014, 42(W1): W252–W258
Case D A, Darden T A, Cheatham T E III. AMBER 12. 2012
Adcock S A, Mccammon J A. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chemical Reviews, 2006, 106(5): 1589–1615
Essmann U, Perera L, Berkowitz M, Darden T, Lee H, Pedersen L G. A smooth particle mesh Ewald method. Journal of Chemical Physics, 1998, 103(19): 8577–8593
Colovos C, Yeates T O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science, 1993, 2(9): 1511–1519
Wiederstein M, Slppl M J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 2007, 35(Web Server): W407–W410
Ercan O, Wels M, Smid E J, Kleerebezem M. Molecular and metabolic adaptations of Lactococcus lactis at near-zero growth rates. Applied and Environmental Microbiology, 2015, 81(1): 320–331
Dijk M V, Bonvin A M. 3D-DART: A DNA structure modeling server. Nucleic Acids Research, 2009, 37(Web Server): W235–W239
Dijk M V, Dijk A D, Hsu V, Boelens R, Bonvin A M. Informationdriven protein-DNA docking using HADDOCK: It is a matter of flexibility. Nucleic Acids Research, 2006, 34(11): 3317–3325
Massova I, Kollman P A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in Drug Discovery and Design, 2000, 18(1): 113–135
Kottalam J, Case D A. Langevin modes of macromolecules: Applications to crambin and DNA hexamers. Biopolymers, 1990, 29(10–11): 1409–1421
Sharp K A, Honig B. Electrostatic interactions in macromolecules: Theory and applications. Annual Review of Biophysics and Biophysical Chemistry, 1990, 19(1): 301–332
Sitkoff D, Sharp K A, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. Journal of Physical Chemistry, 1994, 98(7): 1978–1988
Still W C, Tempczyk A, Hawley R C, Hendirckson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. Journal of the American Chemical Society, 1990, 112 (16): 6127–6129
Acknowledgements
This paper is financially supported by the National Natural Science Foundation of China (Grant No. 31570049).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yuan, L., Wu, H., Zhao, Y. et al. Molecular simulation of the interaction mechanism between CodY protein and DNA in Lactococcus lactis. Front. Chem. Sci. Eng. 13, 133–139 (2019). https://doi.org/10.1007/s11705-018-1737-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11705-018-1737-4