Abstract
Many steppe species reach their (north)westernmost distribution limit in western Central Europe. This also applies to Poa badensis, a rare steppe plant of calcareous rock/sand vegetation. To explore potential differences in reproductive success and genetic composition of peripheral populations, we analysed the absolute (north)westernmost occurrences in Western Germany and populations at the western margin (Eastern Austria) and the centre (Central Hungary) of the Pannonicum, representing a part of the continuous range. Specifically, we discuss the genetic and reproductive constitution of the (north)westernmost exclave and draw conclusions on the species’ biogeographical and conservation history in this region. Therefore, we used two independent molecular marker systems (AFLPs, cpDNA sequences) and a set of performance parameters. Overall, lowest regional genetic diversity was found in Western Germany, which is mainly a result of the specific history of two populations. However, this low genetic diversity was not accompanied by reduced reproductive success. The Eastern Austrian populations showed reduced genetic diversity and predominantly reduced performance, interpreted as a consequence of small population sizes. Central Hungarian populations showed the overall highest genetic diversity and comparatively high performance values. We observed high admixture and haplotype sharing between Austrian and Hungarian populations, indicating gene flow among these regions. In contrast, we interpreted the increased population differentiation within, and the clear distinctiveness of the German exclave as a long-term isolation of these (north)westernmost occurrences. Our results, overall, prove the good constitution of these populations and, together with their particular biogeographical history, highlight their conservation value.
Similar content being viewed by others
Introduction
Investigating the constitution of (rare) species at their absolute distributional periphery, we can test commonly and controversially discussed biogeographical hypotheses, e.g. the ‘abundant centre’ distribution (Sagarin and Gaines 2002) or the ‘central-marginal hypothesis’ (Eckert et al. 2008; see also Sexton et al. 2009; Abeli et al. 2014; Pironon et al. 2016). As a result of more unfavourable conditions at the absolute distribution limit, peripheral populations are generally assumed to have smaller population sizes, occur in lower densities and be genetically more differentiated among each other, compared to equivalents in more central parts of the distribution range. Subsequently, genetic bottlenecks, genetic drift and inbreeding may reduce genetic diversity and influence species’ fitness and potential for adaptation to local conditions. These predictions on the genetic composition were partially detected in several studies on steppe plant species (cf. Pérez-Collazos et al. 2009; Wagner et al. 2011a; but see also Kajtoch et al. 2016). However, few studies have analysed the influence on species’ performance and the expected patterns of reduced reproductive success were mostly not observed (cf. Wagner et al. 2011b; but see also Pironon et al. 2016).
Nevertheless, isolated peripheral populations are not necessarily characterised by such parallel patterns. Besides hypotheses on centrality-marginality phenomena, the existence of extrazonal, small-scale refugia, persisting in situ during the Quaternary or even longer (e.g. Plenk et al. 2017), must also be considered. In Central Europe, where steppe species today reach their absolute westernmost distribution limit (cf. Meusel 1970; Lang 1994), such extrazonal refugia can be found north of the Alps, in the Carpathians and the Pannonian region. Depending on the variability of the ancestral gene pool, as well as the probability of and time for accumulation of de novo mutations and/or effects of lineage sorting (Avise 2000), these relict populations could hold comparatively high levels of unique genetic diversity (cf. Schmitt and Varga 2012; Kajtoch et al. 2016; Plenk et al. 2017). The increasing isolation at the distributional periphery and the subsequently increasing genetic differentiation of populations due to genetic drift may have additionally led to high levels of regional genetic diversity; characteristics similar to those described by Hampe and Petit (2005) for ‘stable’ rear edge populations.
Poa badensis is a perennial, thermophilous grass with a disjunct submediterranean distribution mainly restricted to Central and Southeastern Europe (cf. Figure 1; Meusel et al. 1965), as well as to the southeast of France (Conert 1998). Beside this, there are probably further occurrences in northern Anatolia (cf. Buschmann 1942) and the Caucasus (cf. Conert 1998; Zernov et al. 2015, where P. alpina “incl. P. badensis” is listed). At its (north)westernmost distribution limit the species grows at isolated sites in Central and Western Germany (cf. Blaufuss and Reichert 1992; Hensen and Wesche 2007; Hodvina and Cezanne 2008). These scattered occurrences are considered to be relict localities of a former (postglacially) more widely distributed steppe complex (Meusel 1970; Bredenkamp et al. 2002) and often have a specific conservation history.
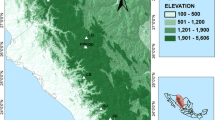
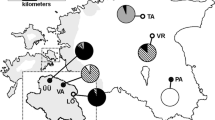
Sampling, distribution and admixture of Poa badensis. Distribution of P. badensis along our transect (a), representing the three study regions (circles) and sampled populations (stars). The species’ continuous Central European distribution is expressed in full-colour, disjunct occurrences as coloured points (distribution map based on Meusel et al. 1965). AFLP-based proportions of admixture (b) derived from Bayesian spatial clustering of individuals (K = 3). For population abbreviations see Table 1
However, the phylogeography of Poa badensis is mostly unknown. The only available genetic study was based on AFLPs and included Central German populations from Saxony-Anhalt and Thuringia, exclusively (Hensen and Wesche 2007). Hensen and Müller (1997) investigated the anemochorous dispersal ability of P. badensis, however, reproductive fitness in wild populations of our study species has not been investigated, previously. Combined studies on performance and genetic characterisation of steppe species are rare (and lacking for P. badensis) and concentrated mostly at a regional scale (cf. Hensen et al. 2005; Dostálek et al. 2010; Lauterbach et al. 2011; Heinicke et al. 2016), with the exception of the work on Stipa capillata by Wagner et al. (2011a, b).
In this study on P. badensis, we focused on the (north)westernmost distribution of this thermophilous (rock) steppe species (cf. Willner et al. 2017) and explored differently peripheral populations within its comparatively narrow Central European distribution range. We included representative populations from the absolute periphery as well as from the margins of its continuous Pannonian distribution range, covering three different study regions. To analyse the genetic structure and diversity of P. badensis along this study transect, we used two molecular marker systems, i.e. Amplified fragment length polymorphisms (AFLPs) and chloroplast (cpDNA) sequence data. We also tested the reproductive success of this rare species across the same populations by estimating different performance parameters in a germination experiment. Based on these datasets, we evaluated the predicted general characteristics of peripheral populations. Specifically, we aimed at answering the following questions and finally draw conclusions on the species’ history in the German exclave: (1) Can we detect different patterns of genetic composition and reproductive success along our study transect, following predictions on centrality-marginality? Or do the patterns rather reflect a particular history of the three study regions, indicating long-term (extrazonal) refugia at the edge of the species’ distribution? (2) Do genetic and performance parameters vary in parallel: are populations of the (north)westernmost exclave less viable, i.e. showing decreased genetic diversity, highly (genetically) isolated populations and reduced reproductive success? Finally (3) we conclude what can be deduced from our results with respect to the species’ biogeographical and conservation history in the (north)westernmost exclave in Central Europe.
Materials and methods
In a statistical large-scale re-classification of Pannonian and Western Pontic steppe grasslands, Willner et al. (2017) defined Poa badensis as a diagnostic species of the order Stipo-Festucetalia pallentis (i.e. rocky steppes; cf. also Borhidi et al. 2012). Accordingly, the species grows on calcareous grus and rock communities, or more rarely on basic substrates, but can also be found on calcareous sand.
P. badensis is wind-pollinated, producing seeds which are dispersed by wind over short distances (Hensen and Müller 1997) and germinate rapidly after release (i.e. with no signs of dormancy; Hensen and Wesche 2007; pers. observ.). The grey-green tussocks produce no vegetative stolons, however, sometimes a clonal reproduction seems to be possible due to tussock fragmentation (cf. Conert 1998, Hensen and Wesche 2007; pers. observ.). Flowering of 4-8 cm long panicles lasts from May to July. One ovate, green to violet spikelet contains 3-6(-9) flowers (Conert 1998). P. badensis is predominantly described as a diploid species (Müntzing and Nygren 1955; Joshi et al. 2017). Nevertheless, the following chromosome numbers have been reported without precise information about the geographical origin: 2n = 14, 28, 42 (Oberdorfer 2001) and 2n = 14, 18, 21, 28 (Conert 2000; including odd numbers). In the Pannonian region in Eastern Austria the species is diploid (Fischer et al. 2008). To test ploidy levels within our dataset, we exemplarily analysed two Austrian and two Hungarian individuals using the CyFlow®Space (Partec, Münster, Germany) and the software FlowMax (version 2.3, Partec 2001). We obtained evidence for a likewise diploid level in these Austrian and Hungarian individuals. In contrast, individuals tested from the German region (n = 4) were observed to be tetraploid (cf. Online Resource 1).
Typically, the ploidy level might be reflected in the number of AFLP fragments, amplified per AFLP phenotype, with higher ploidy levels possibly showing higher fragment numbers (Kardolus et al. 1998). In this sense, the distinctiveness of groups might become blurred by differing averaged AFLP fragment numbers (cf. Guo et al. 2005). However, we observed comparable mean fragment numbers with an even lower value for the assumed tetraploid individuals from the German study region (average 66.4 bands per individual), compared to the assumed diploid individuals from the Pannonian region (average 81.1 bands per individual), and clear grouping patterns in both genetic analyses (i.e. distance-based SplitsTree analysis and Bayesian Clustering analysis; see Results). Therefore, we could reject the possible interference of our AFLP analyses by the difference in ploidy levels and analysed data jointly. Similar un-blurred results were obtained in a number of AFLP studies including differing ploidy levels (e.g. Ma et al. 2010; Španiel et al. 2012).
Study design
Focusing on the Central European distribution of Poa badensis, we defined a study transect (cf. Plenk et al. 2017) crossing Central Hungary and the eastern parts of Lower Austria in the east, and reaching the (north)westernmost isolated occurrences of the species in Western Germany (Fig. 1). More specifically, the Austrian and Hungarian populations are situated at the western margin and in the centre of the Pannonicum, where the species can be found scattered in suitable habitats (Niklfeld 2013; Bartha et al. 2015). In contrast, the German study region in Rhineland-Palatinate (Northern Rhinehesse and Lower Nahe region) nowadays only comprise five currently known populations (Blaufuss and Reichert 1992), which are strongly isolated within this (north)westernmost study region. However, in the past P. badensis was more common in this region, representing (genetically) likely connected occurrences, i.e. a distinct exclave, outside its continuous Pannonian distribution range.
To obtain an equal sampling across our study transect, we chose four populations per study region, i.e. twelve populations in total. In addition to this main dataset, we sampled material from the fifth population known in the Western German study region, the Mainzer Sand (G5). This population was not included in our main dataset due to the low number of individuals (i.e. four) found there, in our sampling year (2010). For molecular analyses, we collected leaf material from randomly chosen individuals across the respective population. Additionally, the number of flowering individuals was estimated at the same time to obtain population-size categories for each sampled population (see Table 1). Seeds of P. badensis were later collected from the same populations (i.e. from 20 individuals per population; again except G5) by taking one, randomly chosen, mature panicle per fruiting plant.
Germination experiment
This experiment was carried out with 20 individuals per population. For each mother plant the number of flowers (shed spikelets were counted to obtain an estimate of the total number of flowers in the panicle) and the number of seeds per panicle were counted. The seeds were distinguished as mature and undeveloped seeds; total seed mass was then determined for all mature seeds per individual panicle. For germination, usually ten seeds per individual mother plant were used, whereby undeveloped, shrunken seeds were excluded. The seeds were placed in petri dishes on a double-layered filter paper following a definite scheme, which allowed tracking each seed, individually. Petri dishes were placed in a climate chamber for 41 days. The settings for light and temperature mimic a day/night cycle with 12 h and 25 °C (with light) and 12 h and 15 °C (dark). The germination process was documented by differentiating four states for each seedling: ‘not germinated’, ‘radicle’ (visible), ‘cotyledon’ (visible), ‘dead’. Individuals, which successfully germinated (i.e. showing breakthrough of the radicle), were transplanted in multi-pot-plates filled with a standardised mixture of plant soil and quartz sand at a ratio of 2 to 1. After 2 months, seedling survival was determined by recording the plants survived in the multi-pot-plates.
Molecular methods
To isolate total genomic DNA from silica dried leaf material, ca. 20 mg per sample were ground with sterilized glass pellets and processed using the DNeasy™ Plant Mini Extraction Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol.
Amplified Fragment Length Polymorphisms (AFLPs) profiles were generated for 7–23 individuals per population according to Vos et al. (1995) with some minor modifications (cf. Kropf et al. 2006; Kropf 2012): DNA was digested using the restriction enzymes MseI and EcoRI. In the subsequent pre-selective amplification, primer pairs each containing one selective nucleotide were used. The selective amplifications were then performed with three primer combinations, with each primer containing two additional selective nucleotides (E + ACG/M + CGG [E37/M57], E + AGA/M + CTG [E39/M61], E + ATG/M + CGG [E45/M57]). These combinations have been applied successfully in previous studies (Kropf et al. 2003; Plenk et al. 2016, 2017) and were labelled by three different fluorescence dyes (E37: NED™, E39: 6-FAM™, E45: HEX™). For genotyping, the PCR products were mixed with an internal size standard (ROX™, ET550-R) and run on a MegaBACE DNA Analysis System (Amersham Biosciences, Freiburg, Germany). The raw data obtained was then aligned with the internal size standard using the program MegaBACE Fragment Profiler 1.2 (Amersham Biosciences) and a peak height threshold of 50 relative fluorescent units (RFUs) within a range of 60–550 base pairs fragment length. The generated presence/absence matrix of AFLP fragments was visually checked to exclude misinterpretations as described by Meudt and Clarke (2007). 54 individuals (i.e. six individuals, different on each plate) were replicated during lab work to test the reproducibility of fragments. Furthermore, blind samples were loaded to check runs for impurities (cf. Bonin et al. 2004).
For cpDNA sequence analyses, two regions, rpL16 intron (primers: rpL16F71, rpL16R1516; Shaw et al. 2005) and atpI-H (primers: atpI, atpH; Shaw et al. 2007) were sequenced for six to nine individuals per population. Both plastid markers have been previously tested for our study species together with other cpDNA markers (i.e. rpL32, rpS16, trnL-trnF) and were selected due to their potential for higher intraspecific variation. The amplification was carried out under standard PCR conditions as described in Plenk et al. (2017). PCR-products (12 µl) were sent to LGC Genomics (Berlin, Germany) for cycle sequencing using the same primers. Alignment of cpDNA sequences obtained was done manually with BioEdit (version 7.2.5, Hall 1999).
Data analyses
Performance parameters
Reproductive success was analysed for one panicle per mother plant using the following parameters: number of flowers, number of seeds, seed mass (g), fruit set (%), germination rate (%), and seedling survival rate (%). The number of flowers per panicle was determined as the total of mature seeds, undeveloped seeds and shed spikelets (i.e. multiplied by inherently six flowers per spikelet). The parameter number of seeds only includes the mature seeds counted in the respective panicle. However, as we must assume that shed spikelets contained mostly mature seeds, this parameter might be underestimated. To assess the magnitude of this possible influence on our results, we calculated the proportion of shed spikelets compared to the total number of spikelets, which was only 17.6%. The number of seeds was also used to calculate a mean weight per seed (i.e. seed mass) from the total seed mass of all mature seeds within a single panicle. Fruit set was calculated as the proportion of mature seeds from the number of flowers whereas the germination rate and seedling survival rate were determined as the proportion of germinated seeds/survived seedlings related to the number of seeds applied in the germination experiment. Since our data does not meet the assumptions of parametric analyses, we performed a non-parametric Kruskal–Wallis rank sum test (Kruskal and Wallis 1952) for regional comparison and a post hoc pairwise multiple comparison between regions (i.e. median differences) following Dunn’s test for stochastic dominance (Dunn 1964; implemented with the R-package ‘dunn.test’; Dinno 2017). Analyses were performed with the statistical program R (version 3.4.0; R Core Team 2017) and a significance level of α = 0.05.
AFLPs
Based on the complementary value of Nei and Li’s similarity coefficient (Nei and Li 1979), we performed an individual-based genetic distance analysis using PAUP* (version 4.0; Swofford 2002). Further, we computed a NeighborNet based on uncorrected P distances using Splits Tree4 (version 4.14.4; Huson and Bryant 2006). Bayesian Clustering was performed with BAPS (version 5.3; Corander et al. 2003, 2004) to reveal the most likely number of clusters (K) by means of stochastic optimisation. Clustering of individuals was performed with a maximum of K = 13 clusters and nine replicates for each K. We performed the analyses with and without geographical coordinates of populations as an informative prior. A Bayesian admixture analysis (Corander and Marttinen 2006) was calculated using the settings described in Plenk et al. (2016). Genetic differentiation among and within populations/regions was estimated in ARLEQUIN (version 3.5.2.2; Excoffier and Lischer 2010) using non-hierarchical and hierarchical analyses of molecular variance (AMOVA; cf. Plenk et al. 2017). These analyses were carried out for the whole dataset and for the (north)westernmost exclave and the Pannonian region, separately. To test for isolation by distance (Wright 1943; Hutchinson and Templeton 1999), pairwise FST-values of the twelve Poa badensis populations were transformed to deal with linear sampling along our study transect (cf. Rousset 1997) and then regressed on geographical distance (i.e. km) among populations. Analyses were performed as described in Plenk et al. (2017) across the study transect and within regional subsets. Genetic (AFLP) diversity was estimated for the studied populations using the program POPGENE (version 1.31; Yeh et al. 1997) and the following diversity indices: Nei’s gene diversity (HE; Nei 1973), Shannon’s information index (I; Shannon and Weaver 1949), and percentage of polymorphic loci (PLP). To reduce potential unbalances caused by the unequal sample size of our AFLP data (ranging from 7–23 individuals per population; Table 1), we additionally carried out a rarefaction analysis for genetic diversity estimation (Petit et al. 1998) using AFLPdiv (version 1.1; Coart et al. 2005). Original AFLP band richness was adjusted to the minimum populational sample size within our dataset (i.e. n = 7). Genetic diversity was then calculated based on the number of AFLP phenotypes (i.e. band richness, Br), which are expected at each locus, for this adjusted sample size. Further, the percentage of polymorphic loci (PLP) was also calculated with this standardised sample size, whereby a locus is considered as polymorphic when Br ranged from 1.05 to 1.95 (5% level) or 1.01 to 1.99 (1% level; Coart et al. 2005).
cpDNA sequence data
Statistical parsimony haplotype networks were created with the program TCS (version 1.21, Clement et al. 2000) based on the combined alignment of both cpDNA sequences. Insertions/deletions which were longer than one base pair, as well as inversions, were coded as single-step mutations; (single) sequence gaps were treated as a fifth character state. Using Arlequin (ver. 3.5.2.2; Excoffier and Lischer 2010) genetic diversity indices, i.e. nucleotide [π] and haplotype diversity [h], were calculated based on cpDNA sequence variation. Testing deviation from neutrality, we further calculated Tajima’s D (Tajima 1989) to detect evidences for possible selection, recent expansion and/or genetic bottlenecks (cf. Christe et al. 2014).
Results
Performance parameters
In total, 1874 seeds from 223 mother plants were included in the germination experiment. On a regional scale, the number of mature seeds per panicle was highest in Central Hungary (mean 38.25 ± 40.28 SD; standard deviation) and lowest in Eastern Austria (mean 22.63 ± 31.05), differing significantly among regions (Kruskal–Wallis test: p = 0.004; Table 2). Results of the post hoc test demonstrated that the number of seeds differed significantly due to the low values observed in the Austrian region. The highest regional fruit set was found in Western Germany (mean 25.8% ± 20.4), whereas Eastern Austrian populations again showed the lowest value (mean 18.6% ± 17.4), differing significantly (Kruskal–Wallis test: p = 0.043). The mean seed mass per seed showed the highest values in the Hungarian (mean 0.3490 mg ± 0.4337) and the lowest in the Eastern Austrian region (mean 0.2899 mg ± 0.1990), demonstrating highly significant differences between regions (p < 0.001). In detail, seed mass was significantly different between the German and the Austrian, as well as the German and the Hungarian study region.
Overall, the level of germination rate after 41 days was high: i.e. 80.8% (± 24.1). The values varied only slightly between 87.9% (± 17.2; G3, Heidesheim) and 71.1% (± 31.0; H1, Budaörs). On regional level, germination was highest in the German exclave (mean 82.8% ± 22.4) and lowest in Central Hungary (mean 77.3% ± 26.8); albeit without statistical significance. A similar pattern was obtained for seedling survival with the regionally highest survival rate in Western Germany (mean 64.3% ± 24.5). The lowest survival rate was found in the Hungarian region (mean 47.5% ± 25.9); here, differences between regions were highly significant (Kruskal–Wallis test: p < 0.001) due to significant differences between the Austrian and Hungarian and the German and Hungarian region. Detailed results of all performance parameters are summarised in Table 2.
None of the performance parameters was significantly correlated with population size, although there was a slight trend to interdependence between fruit set, number of seeds and number of flowers. A significant positive correlation was shown for the seedling survival rate and geographical longitude (r = 0.657, p = 0.02), indicating gradually higher survival rates towards the (north)westernmost exclave. However, none of the other performance parameters was significantly correlated with this estimate of peripherality.
Genetic structure
For each study region about 80 individuals (i.e. 241 in total) were included in the AFLP analyses, whereby for 30 of those samples the generation of reliable AFLP fragment patterns has failed. Therefore, subsequent statistical analyses were based on 211 individuals (Table 1) and 770 polymorphic AFLP fragments (i.e. 259, 217, and 295 of the three primer combinations, respectively; the latter count also including a single monomorphic fragment). In a genetic distance analysis individuals clustered predominantly according to their regional origin, with a high similarity between the Eastern Austrian and Central Hungarian populations and a stronger differentiation to and within the Western German populations. Within regions, grouping of individuals more or less reflected their respective population origin; nevertheless, particularly the population Hundsheimer Berg (A3) showed a heterogeneous composition (cf. Online Resource 2).
Testing the molecular variance (AMOVA) without regional structure, we found variation of 21.8% among and 78.2% within all populations (Online Resource 3). However, using a hierarchical approach based on the geographical origin (i.e. the three study regions), 11.0% of variation originated from the differentiation among regions, whereby 13.1% represented population differentiation within regions. The partitioning of data into two groups, representing the (north)westernmost exclave versus the Pannonian region (i.e. Austrian and Hungarian populations), revealed 12.3% of the variation among these groups and 14.1% among populations within groups. To test patterns of within-region population differentiation, we calculated two additional AMOVA analyses, i.e. one for the (north)westernmost exclave and one for the Pannonian region, resulting in 23.5% and 12.3% population differentiation, respectively (cf. Online Resource 3). The linear regression of pairwise transformed FST-values and geographical distances between populations (i.e. km) revealed a significant isolation-by-distance pattern (Wright 1943) along our study transect (r = 0.4723; p = 0.0037). Within subsets we found significant isolation by distance in the Pannonian region (r = 0.3570; p = 0.0222), but not in the (north)westernmost exclave (r = − 0.2858; p = 0.4628).
Bayesian clustering of individuals revealed an optimal number of K = 3 groups within the whole dataset. Individuals of Austrian and Hungarian populations were again allocated together as one of these three groups, in the following called the Pannonian group/populations (only the populations Hundsheimer Berg, A3 and Csákvár, H4 showed some similarity to the German region). German individuals were differentiated into the two other groups, one containing the populations Eckelsheim (G1) and Weilersberg (G2; i.e. group G I) and the other one Heidesheim (G3) and Eierfels (G4; group G II; data not shown).
The admixture analysis based on spatial clustering of individuals (K = 3; Fig. 1) indicated low gene flow between the (north)westernmost exclave and the Pannonian region: i.e. less admixture in A3, H1, H2 and (almost) no admixture in G1, G2, G3, G4, A1, A2, A4, H3. Only the population Csákvár (H4) showed a comparatively high level of admixture with the German group G I (Fig. 1). The German populations Heidesheim (G3) and Eierfels (G4; i.e. both representing the group G II) demonstrated some admixture with the German group G I (Fig. 1).
Sequences from the chloroplast (regions rpL16, atpI-H) were generated for 94 individuals, resulting in an alignment of 1507 base pairs length and seven haplotypes. We found two main haplotype groups, separated by one specific mutation at bp 883 (atpI-H): one German group and one Pannonian group (i.e. the latter containing individuals exclusively from Austria and Hungary). Within the German group individuals from the population Eckelsheim (G1) formed one specific haplotype, whereas the second haplotype in this group contained all other German populations (i.e. Weilersberg, G2; Heidesheim, G3 and Eierfels, G4). In contrast, the Pannonian group is characterised by further structuring (five haplotypes), but most of the individuals (i.e. 45 of 59 individuals; representatives of all Austrian and Hungarian populations) formed one major haplotype (see Fig. 2).
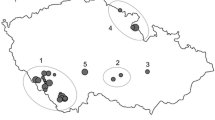
CpDNA haplotype network. TCS haplotype network showing the distinctiveness of the (north)westernmost exclave (G), separated from the Pannonian haplotype group (including Austrian (A) and Hungarian (H) populations) by one specific point mutation (at bp 883, atpI-H; bold lines). The inserted map illustrates the geographical isolation of population G1. For population abbreviations see Table 1
In addition to our main cpDNA dataset, we generated DNA sequences for the Mainzer Sand population (G5). The four individuals analysed were allocated to both haplotypes detected within the German exclave (see Fig. 2 and Online Resource 4).
Genetic diversity
Different genetic diversity estimates based on our AFLP data basically produced similar results (Table 1). Comparing the diversity values (HE) of our studied populations with values compiled by Nybom (2004) for long-lived perennial species (0.25), occurring in a narrow geographical range (0.28), and characterised by an anemochorous seed dispersal (0.27), we found overall a comparatively low absolute genetic diversity in all populations and study regions. The absolute lowest (HE: 0.0457; I: 0.0755) and highest (HE: 0.1408; I: 0.2243) diversity values were found in the populations Eckelsheim (G1, population size category “PSC”: 1) and Pilisszentkereszt (H3, PSC: 4), respectively. Interestingly, the population Weilersberg (G2), showed the second lowest genetic diversity (HE: 0.0479; I: 0.0802), although being a large population (PSC: 4). After standardisation of AFLP band richness to the minimum populational sample size (i.e. rarefaction to 7, cf. Table 1), we revealed adjusted values for genetic diversity (PLP, Br). The estimates after rarefaction were slightly higher in some of the populations (Eckelsheim, G1; Eierfels, G4 and Eggendorf, A2; the latter one being the population with the originally highest number of individuals analysed). However, the highest and lowest values were again found in the populations Pilisszentkereszt (H3) and Eckelsheim (G1), respectively.
On regional average, genetic diversity values (HE, I) were highest in Central Hungary (HE: 0.1122; I: 0.1772) and lowest in Western Germany (HE: 0.0876; I: 0.1398). Genetic diversity of Eastern Austrian populations was only slightly higher (HE: 0.0906; I: 0.1438) than in the (north)westernmost exclave. However, the distribution of genetic diversity across regions varied slightly for PLPs: here, the lowest regional diversity was found in the Eastern Austrian study region and this pattern retained also after rarefaction.
Genetic diversity calculated from the main cpDNA sequence data showed overall very low diversity in the German region as each population (excluding G5) was monomorphic (Table 1). The highest population diversity was found in the populations Hundsheimer Berg (A3; haplotype diversity: 0.5238) and Csákvár (H4; nucleotide diversity: 0.0006). At the regional level, cpDNA haplotype diversity of the Austrian and Hungarian study region was similar (i.e. 0.40475 for Austria and 0.38095 for Hungary), while the Hungarian region showed the highest nucleotide diversity. Testing deviation from neutrality (i.e. Tajima’s D; Tajima, 1989) revealed no significant indications for departure from neutrality. None of the genetic diversity estimates was correlated with population size, geographical longitude (i.e. peripherality) or one of the performance parameters.
Discussion
Species’ performance and genetic composition along the study transect
By testing biogeographical hypotheses, e.g. a central-marginal gradient, we evaluated demographic characteristics (i.e. population density, population size), reproductive success and genetic composition along our study transect. Regarding the demographic characterisation of our studied populations, Poa badensis followed the hypothesised gradient (Sagarin and Gaines 2002; Abeli et al. 2014). In Germany, the species is classified as ‘vulnerable’ and regionally ‘endangered’, e.g. in Rhineland-Palatinate where only a few populations exist (see chapters below; Korneck et al. 1996). Although we have no concrete information on the total number of existing populations in Austria and Hungary, the species is more common there (Niklfeld 2013; Bartha et al. 2015). Hence, we conclude that the density of populations in our study regions is indeed declining towards the species (north)westernmost distribution limit. Moreover, we analysed the individual-richest populations in Hungary, represented by hundreds of individuals. The smallest populations within our study transect, most contained less than 200 individuals (Tables 1, 2), were found in Eastern Austria. However, the individual-poorest population analysed was again from Western Germany (i.e. Eckelsheim, n < 30).
On the contrary, our results did not indicate the expected reduced reproductive success of the (north)westernmost populations (cf. Abeli et al. 2014). The values of performance parameters were highest or at least equally high as values from the continuous part of the distribution range (i.e. Central Hungary; Table 2), as also described for Stipa capillata (Wagner et al. 2011b) and Lychnis viscaria (Lammi et al. 1999). Nevertheless, our performance parameters include some uncertainty, since they reflect the species’ reproductive success in situ (i.e. in the wild; cf. Wagner et al. 2011b) and may therefore include some maternal effects (Gutterman 2000).
Regarding the genetic composition of our study species, we observed a regional genetic impoverishment of (but see next Discussion chapter) and an increasing genetic differentiation among the (north)westernmost populations, as expected for a central-marginal gradient (Sagarin and Gaines 2002; Eckert et al. 2008; Sexton et al. 2009), but also partially for ‘stable’ rear edge populations (rather than for leading edge populations; Hampe and Petit 2005). Regional population differentiation was by far the highest in Germany. Pannonian populations from Austria and Hungary showed a lower population differentiation (Online Resource 3), which should also be found when further populations from the centre of the continuous Pannonian range are analysed, because gene flow is obviously high in this region. Both marker systems (AFLP & cpDNA) revealed a clear distinctiveness of the (north)westernmost exclave. Such a strong genetic distinctiveness and/or high population differentiation of peripheral populations has been observed, for example, in two other Poaceae species, Stipa capillata (Hensen et al. 2010; Wagner et al. 2011a) and Stipa pennata (Wagner et al. 2012), as well as in our previous study on Linum flavum (Plenk et al. 2017), which is based on a similar transect setting.
Constitution of populations in the (north)westernmost isolated exclave
In the German exclave, the species’ reproductive success was not obviously reduced, while at the same time our analyses of genetic composition showed a partial genetic impoverishment. This pattern was somewhat surprising, as we did not expect to find reduced genetic diversity and high reproductive success in this region. The overall low level of genetic diversity observed in Western Germany, seems to be mainly a result of very low values found in two of the four investigated populations (i.e. Eckelsheim, G1, PLP % [7]: 21.7; Weilersberg, G2, PLP % [7]: 24.0). The populations Heidesheim (G3, PLP % [7]: 49.7) and Eierfels (G4, PLP % [7]: 44.7) did not show reduced levels of genetic diversity; the values observed there are comparable to those of the continuous distribution in Central Hungary (PLP % [7]: 41.1; see Table 1). Hensen and Wesche (2007) observed comparatively high levels of genetic diversity (PLP %: 50.0–74.2; genetic diversity: 0.20–0.29) in Central German populations of Poa badensis, but a moderate to low genetic differentiation among populations. Compared to other thermophilic steppe species from the same plant family (i.e. Stipa capillata, Hensen et al. 2010; Stipa pennata agg., Wagner et al. 2012; Durka et al. 2013), or other steppe species occurring in calcareous rock/sand vegetation (i.e. Globularia bisnagarica, Honnay et al. 2007; Alyssum montanum, Španiel et al. 2012), these values represent high levels of genetic diversity at the (north)westernmost border of their distribution. Consequently, Hensen and Wesche (2007) described the species as only slightly affected by fragmentation and assessed genetic exchange between populations in their study region as still possible. In our more western German study region, P. badensis is obviously affected by reduced gene flow (cf. Figures 1, 2), which is also clearly demonstrated by the strong genetic within-region population differentiation (Online Resource 3). Although populations of wind-pollinated and -dispersed species in general are assumed to have a greater potential for genetic exchange, the low dispersal capacity of our study species together with the scattered availability of suitable habitats in our study region have led to this increased genetic differentiation. In addition, we found no clear evidence for isolation by distance, possibly indicating genetic drift already occurring in the Western German populations.
Even though the regional genetic diversity is lowest in the German exclave, we basically interpret its comparatively high reproductive success as indicator for good reproductive capability. As we obtained indications of different ploidy levels within our study transect (i.e. diploid populations in Austria and Hungary, but tetraploid populations in Germany, Online Resource 1), a possible explanation for the high performance parameters observed in the (north)westernmost exclave might be the occurrence of one (or more) polyploidization events, likely occurring during warm (postglacial) stages. This may have led to a stronger reproductive potential, probably as an evolutionary strategy to overcome the more unfavorable conditions at the absolute range limit. Moreover, polyploid populations may also show generally higher levels of genetic diversity (cf. Alyssum montanum, Španiel et al. 2011).
However, the species’ biogeographical history and specific traits (e.g. longevity) may have aided to retain a certain level of genetic diversity, as observed in the populations Heidesheim (G3) and Eierfels (G4). On the contrary, the populations Eckelsheim (G1) and Weilersberg (G2) are likely less viable and show low genetic diversity, which is driven by their histories. Although population size did not correlate with the genetic diversity in general, at least one of these two populations is characterised by small population size (G1, less than 30 individuals). Nevertheless, low genetic diversity of populations does not necessarily imply that they have to be less viable. Habel and Schmitt (2012, 2018) argued that specialised species, as a consequence of habitat loss and degradation, are commonly characterised by such low levels of genetic diversity accompanied by high genetic distinctiveness. However, they may persist in small-sized remnants of suitable habitats over long periods of time, just slightly affected by population bottlenecks and inbreeding due to their limited, but locally adapted genetic composition.
Regional biogeography and conservation history of the German exclave
Ellenberg and Leuschner (2010) quote Poa badensis among other steppe species as a relict, surviving on sandy/rocky grassland sites in Central Europe since the last glacial. However, they also remarked that most of the populations occurring today are likely younger (i.e. originating from Neolithic Age or early mediaeval times; cf. Willis et al. 1998). In Germany, relict localities of thermophilic species are described from dry regions like the Unstruttal (Becker 1999), the (Lower) Nahe region, aeolian deposits of large rivers (e.g. the north and east of the Rhinehessian plateau; Blaufuss and Reichert 1992; Witschel 1991) or south-exposed rock faces of low mountain ranges (e.g. the Swabian Alb; Wilmanns 2005). Our German study region is situated in two of these refugial areas, the Rhinehessian plateau and the adjacent Lower Nahe region. They are characterised by calcareous aeolian sands, deposited along the northern slopes of the plateau in glacial and early postglacial times (Jännicke 1892; Ellenberg and Leuschner 2010), as well as by the basic conglomerate of the so-called Waderner Schichten of the Upper Rotliegend Group (Weiss 1889).
In our study, we included four of the five mentioned populations from Rhineland-Palatinate by performing all analyses, which overall characterised the (north)westernmost exclave as genetically impoverished. However, there are two populations, Eierfels (G4) and Heidesheim (G3), showing comparatively high levels of genetic diversity. As their history is very different, we will go into more detail. The population Eierfels (G4) is situated on a flat ridge of basic conglomerate in the Lower Nahe region which is protected as natural monument and harbours a unique xerothermic vegetation (Korneck 1998; Oesau and Frahm 2007). The population of P. badensis is rather small, but likely represents an old occurrence probably existing there since the early postglacial (cf. Blaufuss and Reichert 1992 and citation therein). In Heidesheim (G3) our study species grows secondary on a cemetery wall. This rather large and likewise old population persists there over a long period of time and represents one of the two still existing populations on the northern edge of the Rhinehessian plateau (i.e. beside the relict population in the Mainzer Sand). It seems possible that the species had arrived there during the construction of the wall with limestone mortar or, shortly after completion, via natural dispersal (e.g. by wind) from the close surrounding area, where the species previously occurred more frequently (cf. Blaufuss and Reichert 1992; Hodvina and Cezanne 2008).
Occurring in the close vicinity of Heidesheim (i.e. only 1.5 km apart), the population Weilersberg (G2) is characterised by a different and considerably younger history. This area was originally used as a sandpit and later designated as a nature reserve. To our knowledge, P. badensis had been introduced to this site by taking (seed) material from the closely located population Heidesheim (G3). However, in our AFLP data we found the population Weilersberg grouping together with Eckelsheim and not with the assumed source of the seed transfer (Fig. 1). Therefore, we may assume that possibly a second gene pool has been used during the introduction, or that based on a heterogeneous source, lineages were sorted out differently given population bottlenecks. Considering this specific history, the extremely low genetic diversity observed in this large population is not surprising and likely reflects this artificial (recent) founder event. However, the reproductive success in general was not significantly reduced by this bottleneck. Geographically most isolated from the other occurrences of P. badensis in Western Germany, the population Eckelsheim (G1) is again located on a rocky outcrop belonging to the Waderner Schichten as part of the Rhinehessian Plateau (Fig. 1). This isolated location is also reflected in our cpDNA results, where Eckelsheim represents one distinct haplotype within the German haplotype group (Fig. 2). The estimated genetic diversity of this population was by far the absolute lowest from all analysed populations, which we attribute to the obvious lack of gene flow and the very low population size (Table 1). Nevertheless, in combination with the not reduced reproductive success of this population, an adaptation to low genetic diversity as described by Habel and Schmitt (2012; 2018) might be assumed.
Our AFLP data revealed admixture between the populations Heidesheim (G3) and Eierfels (G4; together representing the group GII, Fig. 1) and the German group GI. Due to small population size (Eckelsheim, G1) and recent translocation (Weilersberg, G2) the other two German populations missed such an admixture pattern. To a lesser extent we also found admixture between the German group GI and the Pannonian region, indicating an earlier genetic connection of the (north)westernmost exclave with the continuous Pannonian occurrences. Despite that, we observed that 123 (30.4%) of 404 regionally unique AFLP fragments (cf. Petit et al. 1998) were only present in the (north)westernmost exclave. Within the continuous Pannonian distribution area, 281 (69.6%) of these regionally unique AFLP fragments were found. In this context, having nearly one-third of unique AFLP fragments, the peripheral populations in Western Germany represent a considerable and unique part of the total species’ genetic variation (cf. Plenk et al. 2017). Moreover, we obtained evidence for the relict status and high conservation relevance of the Mainzer Sand population (G5) although it was represented by just four individual plants. The four individuals analysed were allocated to both detected haplotypes of the German exclave (cf. Figure 2 and Online Resource 4), demonstrating that this probably oldest relict population (Korneck and Pretscher 1984) might still hold the full spectrum of genetic diversity original to this region.
Therefore, we interpret the observed genetic composition, demographic traits and reproductive success within the German exclave as evidence of a long-term (successful) in situ survival, probably since early postglacial times, which we especially hypothesise for the populations Eckelsheim (G1), Eierfels (G4), and Mainzer Sand (G5). We moreover conclude, that the populations of the Waderner Schichten (e.g. Eierfels, Eckelsheim) may originate from the probably slightly earlier populated occurrences along the northern edge of the Rhinehessian plateau (e.g. Mainzer Sand and other previously existing ones) and spread there along the river systems of Nahe and subsidiary streams to the west and south (cf. Blaufuss and Reichert 1992; Hodvina and Cezanne 2008).
Nevertheless, we also have to consider that the species may have reached suitable sites in Western Germany more recently via a long-distance dispersal (LDD) event. In this case, we would expect low genetic diversity and less genetic differentiation of these founder populations from a potential source in more central/eastern areas (cf. Wróblewska 2008). Although we observed some admixture between the German and Pannonian region, our data clearly showed a genetic distinctiveness of the (north)westernmost exclave in both marker systems (cf. Figures 1, 2). We, therefore, concluded that the populations of the German exclave do not represent descendants of a recent LDD-event. However, analysing additional populations of P. badensis from the few existing occurrences “linking” the German exclave and the Pannonian region may strengthen this conclusion.
Due to their rarity, peripheral populations are often under protection, although they are generally considered as genetically impoverished and less competitive (Lesica and Allendorf 1995). With our study on P. badensis, we could clearly demonstrate that these assumptions are not applicable in general. However, due to the low abundance and the lack of connectivity, populations might be nevertheless under pressure if they are not (yet) adapted to low levels of genetic diversity (Habel and Schmitt 2012; 2018). Therefore, we must also consider a possible time lag (Lindborg and Eriksson 2004) between the decline of population sizes and the species’ reproductive success and genetic diversity (cf. Kropf 2012).
References
Abeli T, Gentili R, Mondoni A et al (2014) Effects of marginality on plant population performance. J Biogeogr 41:239–249
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Bartha D, Király G, Schmidt D et al (eds) (2015) Magyarország edényes növényfajainak elterjedési atlasza/Distribution atlas of vascular plants of Hungary. Nyugat-magyarországi Egyetem Kiadó/University of West Hungary Press, Sopron
Becker T (1999) Die Xerothermrasen-Gesellschaften des unteren Unstruttales und einige ökologische Gründe für ihre Verteilung im Raum. Mitt Florist Kartierung Sachsen-Anhalt 4:3–29
Blaufuss A, Reichert H (1992) Die Flora des Nahegebietes und Rheinhessen. Pollichia, Bad Dürkheim
Bonin A, Bellemain E, Bronken Eidesen P et al (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Borhidi A, Kevey B, Lendvai G (2012) Plant communities of Hungary. Akadémiai Kiadó, Budapest
Bredenkamp GJ, Spada F, Kazmierczak E (2002) On the origin of northern and southern hemisphere grasslands. Plant Ecol 163:209–229
Buschmann A (1942) Zur Klärung des Formenkreises um Poa badensis HAENKE. Österreichische Bot Zeitschrift 91:81–130
Christe C, Caetano S, Aeschimann D et al (2014) The intraspecific genetic variability of siliceous and calcareous Gentiana species is shaped by contrasting demographic and re-colonization processes. Mol Phylogenet Evol 70:323–336
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Coart E, Van Glabeke S, Petit RJ et al (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conserv Genet 6:259–273
Conert HJ (1998) Poa. In: Conert HJ (ed) Gustav Hegi, Illustrierte Flora von Mitteleuropa. Band 1. Teil 3. Spermatophyta: Angiospermae: Monocotyledones 1(2). Poaceae (Echte Gräser oder Süßgräser), 3rd revise. Parey Buchverlag, Berlin, p 898
Conert HJ (2000) Pareys Gräserbuch. Die Gräser Deutschlands erkennen und bestimmen. Parey, Berlin
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Corander J, Waldmann P, Sillanpää MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Corander J, Waldmann P, Marttinen P, Sillanpää MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20:2363–2369
Dinno A (2017) dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. R-package
Dostálek T, Münzbergová Z, Plačková I (2010) Genetic diversity and its effect on fitness in an endangered plant species, Dracocephalum austriacum L. Conserv Genet 11:773–783
Dunn OJ (1964) Multiple comparisons using rank sums. Technometrics 6:241–252
Durka W, Nossol C, Welk E et al (2013) Extreme genetic depauperation and differentiation of both populations and species in Eurasian feather grasses (Stipa). Plant Syst Evol 299:259–269
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Ellenberg H, Leuschner C (2010) Vegetation Mitteleuropas mit den Alpen in ökologischer, dynamischer und historischer Sicht, 6th edn. Verlag Eugen Ulmer, Stuttgart
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fischer MA, Oswald K, Adler W (2008) Exkursionsflora für Österreich, Liechtenstein und Südtirol, 3rd edn. Biologiezentrum der Oberösterreichischen Landesmuseen, Linz
Guo Y-P, Saukel J, Mittermayr R, Ehrendorfer F (2005) AFLP analyses demonstrate genetic divergence, hybridization, and multiple polyploidization in the evolution of Achillea (Asteraceae-Anthemideae). New Phytol 166:273–290
Gutterman Y (2000) Maternal effects on seeds during development. In: Fenner M (ed) Seeds: the ecology of regeneration in plant communities, 2nd edn. CABI Publishing, Wallingford, pp 59–84
Habel JC, Schmitt T (2012) The burden of genetic diversity. Biol Conserv 147:270–274
Habel JC, Schmitt T (2018) Vanishing of the common species: empty habitats and the role of genetic diversity. Biol Conserv 218:211–216
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Heinicke S, Hensen I, Rosche C et al (2016) Fragmentation and environmental constraints influence genetic diversity and germination of Stipa pennata in natural steppes. Flora 224:42–49
Hensen I, Müller C (1997) Experimental and structural investigations of anemochorous dispersal. Plant Ecol 133:169–180
Hensen I, Wesche K (2007) Genetic structure of the rare Poa badensis (Poaceae) in Central Germany. Nov Hedwigia Supplement 131:177–186
Hensen I, Oberprieler C, Wesche K (2005) Genetic structure, population size, and seed production of Pulsatilla vulgaris Mill. (Ranunculaceae) in Central Germany. Flora 200:3–14
Hensen I, Kilian C, Wagner V et al (2010) Low genetic variability and strong differentiation among isolated populations of the rare steppe grass Stipa capillata L. in Central Europe. Plant Biol 12:526–536
Hodvina S, Cezanne R (2008) Das Badener Rispengras (Poa badensis) in Hessen. Bot und Naturschutz Hessen 21:33–60
Honnay O, Adriaens D, Coart E et al (2007) Genetic diversity within and between remnant populations of the endangered calcareous grassland plant Globularia bisnagarica L. Conserv Genet 8:293–303
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hutchinson DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Jännicke W (1892) Die Sandflora von Mainz, ein Relict aus der Steppenzeit. Gebrüder Knauer, Frankfurt am Main
Joshi A, Bushmann S, Pickett B et al (2017) Phylogenetic relationships among low ploidy Poa species using chloroplast sequences. Genome 60:384–392
Kajtoch Ł, Cieślak E, Varga Z et al (2016) Phylogeographic patterns of steppe species in Eastern Central Europe: a review and the implications for conservation. Biodivers Conserv 25:2309–2339
Kardolus JP, Van Eck HJ, Van Den Berg RG (1998) The potential of AFLPs in biosystematics: a first application in Solanum taxonomy (Solanaceae). Plant Syst Evol 210:87–103
Korneck D (1998) Carex halleriana im Nahegebiet (zweiter Bericht). Decheniana 151:41–49
Korneck D, Pretscher P (1984) Pflanzengesellschaften des Naturschutzgebietes “Mainzer Sand” und Probleme ihrer Erhaltung. Natur und Landschaft 59:307–315
Korneck D, Schnittler M, Vollmer I (1996) Rote Liste der Farn- und Blütenpflanzen (Pteridophyta et Spermatophyta) Deutschlands. In: Ludwig G, Schnittler M (eds) Rote Liste gefährdeter Pflanzen Deutschlands, Schriftenr. Bundesamt für Naturschutz, Bonn-Bad Godesberg, pp 21–187
Kropf M (2012) Genetic variation, biogeographical history, and conservation of Anthyllis montana L. ssp. jacquinii (Kern.) Hayek (Fabaceae) at its northern distribution limit. Int J Plant Sci 173:789–801
Kropf M, Kadereit JW, Comes HP (2003) Differential cycles of range contraction and expansion in European high mountain plants during the Late Quaternary: insights from Pritzelago alpina (L.) O. Kuntze (Brassicaceae). Mol Ecol 12:931–949
Kropf M, Comes HP, Kadereit JW (2006) Long-distance dispersal vs vicariance: the origin and genetic diversity of alpine plants in the Spanish Sierra Nevada. New Phytol 172:169–184
Kruskal WH, Wallis A (1952) Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621
Lammi A, Siikamäki P, Mustajärvi K (1999) Genetic diversity, population size, and fitness in central and peripheral populations of a rare plant Lychnis viscaria. Conserv Biol 13:1069–1078
Lang G (1994) Quartäre Vegetationsgeschichte Europas. Gustav Fischer Verlag, Jena, New York
Lauterbach D, Ristow M, Gemeinholzer B (2011) Population genetics and fitness in fragmented populations of the dioecious and endangered Silene otites (Caryophyllaceae). Plant Syst Evol 298:155–164
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760
Lindborg R, Eriksson O (2004) Historical landscape connectivity affects present plant species diversity. Ecology 85:1840–1845
Ma JX, Li YN, Vogl C et al (2010) Allopolyploid speciation and ongoing backcrossing between diploid progenitor and tetraploid progeny lineages in the Achillea millefolium species complex: analyses of single-copy nuclear genes and genomic AFLP. BMC Evol Biol 10:11
Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci 12:106–117
Meusel H (1970) Verbreitungsgrenzen südlicher Florenelemente in Mitteldeutschland. Feddes Repert 81:289–309
Meusel H, Jäger E, Weinert E (1965) Vergleichende Chorologie der zentraleuropäischen Flora, vol I. VEB Gustav FischerVerlag, Jena
Müntzing A, Nygren A (1955) A new diploid variety of Poa alpina with two accessory chromosomes at meiosis. Hereditas 41:405–422
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Niklfeld H (2013) Arealmuster von Pflanzen der Xerothermrasen und ihrer Kontaktgesellschaften im Raum von Wien (Österreich). In: Thüringer Ministerium für Landwirtschaft Forsten Umwelt und Naturschutz (ed) Steppenlebensräume Europas - Gefährdung, Erhaltungsmassnahmen und Schutz. Thüringen, pp 39–42
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Oberdorfer E (2001) Pflanzensoziologische Exkursionsflora für Deutschland und angrenzende Gebiete, 8th edn. Ulmer, Stuttgart
Oesau A, Frahm J-P (2007) Führer zu bryologischen Exkursionen. Der Eierfels bei Dorsheim. Arch Bryol 23:1–6
Partec (2001) FlowMax software for cytometry, operating manual—data analysis. Partec GmbH, Münster, Germany
Pérez-Collazos E, Sanchez-Gómez P, Jiménez F, Catalán P (2009) The phylogeographical history of the Iberian steppe plant Ferula loscosii (Apiaceae): a test of the abundant-centre hypothesis. Mol Ecol 18:848–861
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pironon S, Papuga G, Villellas J et al (2016) Geographic variation in genetic and demographic performance: new insights from an old biogeographical paradigm. Biol Rev. https://doi.org/10.1111/brv.12313
Plenk K, Göd F, Kriechbaum M, Kropf M (2016) Genetic and reproductive characterisation of seasonal flowering morphs of Gentianella bohemica revealed strong reproductive isolation and possible single origin. Plant Biol 18:111–123
Plenk K, Bardy K, Höhn M et al (2017) No obvious genetic erosion, but evident relict status at the westernmost range edge of the Pontic-Pannonian steppe plant Linum flavum L. (Linaceae) in Central Europe. Ecol Evol 7:6527–6539
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved from https://www.r-project.org
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Sagarin RD, Gaines SD (2002) The “abundant centre” distribution: to what extent is it a biogeographical rule? Ecol Lett 5:137–147
Schmitt T, Varga Z (2012) Extra-Mediterranean refugia: the rule and not the exception? Front Zool 9:22
Sexton JP, McIntyre PJ, Angert AL, Rice KJ (2009) Evolution and ecology of species range limits. Annu Rev Ecol Evol Syst 40:415–436
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press, Urbana
Shaw J, Lickey EB, Beck JT et al (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Španiel S, Marhold K, Filová B, Zozomová-Lihová J (2011) Genetic and morphological variation in the diploid-polyploid Alyssum montanum in Central Europe: taxonomic and evolutionary considerations. Plant Syst Evol 294:1–25
Španiel S, Marhold K, Thiv M, Zozomová-Lihová J (2012) A new circumscription of Alyssum montanum ssp. montanum and A. montanum ssp. gmelinii (Brassicaceae) in Central Europe: molecular and morphological evidence. Bot J Linn Soc 169:378–402
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. 142
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Vos P, Hogers R, Bleeker M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wagner V, Durka W, Hensen I (2011a) Increased genetic differentiation but no reduced genetic diversity in peripheral vs. central populations of a steppe grass. Am J Bot 98:1173–1179
Wagner V, von Wehrden H, Wesche K et al (2011b) Similar performance in central and range-edge populations of a Eurasian steppe grass under different climate and soil pH regimes. Ecography 34:498–506
Wagner V, Treiber J, Danihelka J et al (2012) Declining genetic diversity and increasing genetic isolation toward the range periphery of Stipa pennata, a Eurasian feather grass. Int J Plant Sci 173:802–811
Weiss E (1889) Die Gliederung des Rothliegenden im Saar-Nahe-Gebiet. In: Grebe H (ed) Erläuterung zu Blatt Losheim. Wadern und Wahlen der geologischen Spezialkarte von Preussen, Berlin
Willis KJ, Sümegi P, Braun M et al (1998) Prehistoric land degradation in Hungary: who, how and why? Antiquity 72:101–113
Willner W, Kuzemko A, Dengler J et al (2017) A higher-level classification of the Pannonian and western Pontic steppe grasslands (Central and Eastern Europe). Appl Veg Sci 20:143–158
Wilmanns O (2005) Kältezeitliche Reliktpflanzen der Schwäbischen Alb Aktualisierte Überlegungen zur prähistorischen Landschaft. Hoppea, Denkschrift der Regensburgischen Bot Gesellschaft 66:447–468
Witschel M (1991) Die Trinia glauca-reichen Trockenrasen in Deutschland und ihre Entwicklung seit 1880. Berichte der Bayer Bot Gesellschaft 62:189–220
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wróblewska A (2008) From the center to the margins of geographical range: molecular history of steppe plant Iris aphylla L. in Europe. Plant Syst Evol 272:49–65
Yeh FC, Yang R-C, Boyle TBJ et al (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Alberta
Zernov AS, Alekseev YE, Onipchenko VG (2015) Flora of vascular plants of the Karachay-Cherkess Republic (Oпpeдeлитeль cocyдиcтыx pacтeний Кapaчaeвo-Чepкeccкoй Pecпyблики). KMK Scientific Press, Moscow
Acknowledgements
Open access funding provided by Austrian Science Fund (FWF). We would like to thank the FWF—Austrian Science Funds for granting two steppe plant projects: P21951-B16 to MK and P27955-B29 to Wolfgang Willner (University of Vienna) and to MK. Specifically, we thank Isabella Auberger and Martin Bramböck for their support during the germination experiment and Silviya Wolkerstorfer, Gudrun Kohl and the lab-team of restoration ecology at the TU Munich for support in the DNA lab. We would like to thank all people involved in collecting plant material as well as the authorities providing us respective permits. For the careful reading of our manuscript, we especially thank our colleague Monika Kriechbaum as well as David Horner for improving the English. Moreover, we thank Thomas Schmitt (Müncheberg) and one anonymous reviewer for their constructive comments on our manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Daniel Sanchez Mata.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Plenk, K., Bardy, K., Höhn, M. et al. Long-term survival and successful conservation? Low genetic diversity but no evidence for reduced reproductive success at the north-westernmost range edge of Poa badensis (Poaceae) in Central Europe. Biodivers Conserv 28, 1245–1265 (2019). https://doi.org/10.1007/s10531-019-01722-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-019-01722-x