Abstract
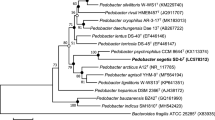
A Gram-stain-negative, aerobic, non-motile, rod-shaped and non-spore-forming bacterium, designated YXT, was isolated from wetland soil. Compared to strain YXT, Pedobacter yonginense HMD1002T had the highest 16S rRNA gene sequence identity (97.8%), and the remaining strains had the identities below 97%. Phylogenetic analysis of the 16S rRNA gene sequences showed that strain YXT grouped with P. yonginense HMD1002T. The values of DNA–DNA hybridization and genomic orthologous average nucleotide identity (orthoANI) between strain YXT and Pedobacter yonginense KCTC 22721T were 58.5% and 82.0%, respectively. The genome size of strain YXT was 5.0 Mb, comprising 4369 predicted genes with a DNA G+C content of 37.3 mol %. Strain YXT had menaquinone-7 as the only respiratory quinone and polar lipids of phosphatidylethanolamine, sphingolipid, aminolipid and three unidentified lipids. The predominant cellular fatty acids (> 10%) of strain YXT were summed feature 3 (iso-C15:0 2-OH and/or C16:1ω7c), iso-C15:0 and iso-C17:0 3-OH. Strain YXT could be distinguished from the other Pedobacter members based on the data of phylogenetic distance, DNA–DNA hybridization, genomic orthoANI, RecA MLSA, core-protein comparison, and hydrolyses of l-arginine, utilization of d/l-lactate, l-alanine, 5-ketonic gluconate and glycogen. Therefore, strain YXT represents a novel species of the genus Pedobacter, for which the name Pedobacter paludis sp. nov. is proposed. The type strain is YXT (= KCTC 62520T = CCTCC AB 2018029T).


Similar content being viewed by others
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Collins MD (1985) Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Cowan ST, Steel KJ (1970) Manual for the identification of medical bacteria. Q Rev Biol 17
Da X, Jiang F, Chang X, Ren L, Qiu X, Kan W, Zhang Y, Deng S, Fang C, Peng F (2015) Pedobacter ardleyensis sp. nov., isolated from soil in Antarctica. Int J Syst Evol Microbiol 65:3841–3846
Dahal RH, Kim J (2016) Pedobacter humicola sp. nov., a member of the genus Pedobacter isolated from soil. Int J Syst Evol Microbiol 66:2205–2211
David M, Emms S, Kelly (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157
De Ley J, Cattoir H, Reynaerts A (1970)) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Du J, Singh H, Ngo HT, Won KH, Kim KY, Yi TH (2015) Pedobacter daejeonensis sp. nov. and Pedobacter trunci sp. nov., isolated from an ancient tree trunk, and emended description of the genus Pedobacter. Int J Syst Evol Microbiol 65:1241–1246
Fautz E, Reichenbach H (1980) A simple test for flexirubin-typepigments. FEMS Microbiol Lett 8:87–91
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Mol Biol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinform 35(1):6.12.1–6.12.19
Fitch WM (1971) Toward defining the course of evolution: minimumchange for a tree topology. Syst Zool 20:406–416
Gao JL, Sun P, Mao XJ, Du YL, Liu BY, Sun JG (2017) Pedobacter zeae sp. nov., an endophytic bacterium isolated from maize root. Int J Syst Evol Microbiol 67:231–236
Gordon NS, Valenzuela A, Adams SM, Ramsey PW, Pollock JL, Holben WE, Gannon JE (2009) Pedobacter nyackensis sp. nov., Pedobacter alluvionis sp. nov. and Pedobacter borealis sp. nov., isolated from Montana flood-plain sediment and forest soil. Int J Syst Evol Microbiol 59:1720–1726
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Hugh R, Leifson E (1953) The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J Bacteriol 66:24–26
Hwang CY, Choi DH, Cho BC (2006) Pedobacter roseus sp. nov., isolated from a hypertrophic pond, and emended description of the genus Pedobacter. Int J Syst Evol Microbiol 56:1831–1836
Joung Y, Kim H, Joh K (2010) Pedobacter yonginense sp. nov., isolated from a mesotrophic artificial Lake in Korea. J Microbiol 48:536–540
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal-X version 2.0. Bioinformatics 23:2947–2948
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39(D):225–229
Marmur J, Schildkraut CL, Doty P (1961) The reversible denaturation of DNA and its use in studies of nucleic acid homologies and the biological relatedness of microorganisms. J Chim Phys Physicochim Biol 58:945–955
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by highperformance liquid chromatography. Int J Syst Bacteriol 39:159–167
Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:182–185
Prescott LM, Harley JP (2001) The effects of chemical agents on bacteria II: antimicrobial agents (Kirby–Bauer Method). In: Prescott LM, Harley P (eds) Laboratory exercises in microbiology. McGraw-Hill, New York, pp 257–262
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. In: MIDI Technical Note. MIDI Inc, Newark
Sawabe T, Kita-Tsukamoto K, Thompson FL (2007) Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol 189:7932–7936
Steyn PL, Segers P, Vancanneyt M, Sandra P, Kersters K, Joubert JJ (1998) Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. proposal of the family Sphingobact. Int J Syst Bacteriol 48:165–177
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tarrand JJ, Gröschel DH (1982) Rapid, modified oxidase test for oxidase-variable bacterial isolates. J Clin Microbiol 16:772–774
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Wang B, Li X, Zheng F, Liu R, Quan J, Liang H, Guo S, Guo G, Zhang J (2007) pEGFP-T, a novel T-vector for the direct, unidirectional cloning and analysis of PCR-amplified promoters. Biotechnol Lett 29:309–312
Weisburg WG, Barns SM, Pelletier BA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wu S, Zhu ZW, Fu L, Li W (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genom 12:444
Yoon JH, Kang SJ, Oh TK (2007) Pedobacter terrae sp. nov., isolated from soil. Int J Syst Evol Microbiol 57:2462–2466
Yoon SH, Sm H, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Acknowledgements
We are grateful to Professor Prof. Bernhard Schink for the Latin construction of the species name. Fatty acids and polar lipid analyses were performed by Guangdong Detection Centre of Microbiology, P. R. China. This work was financially supported by the National Key Research and Development Program of China (2016YFD0800702) and National Natural Science Foundation of China (31870086).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
The GenBank/EMBL/DDBJ accession number of the 16S rRNA gene sequence of strain YXT is MH307976. The genome numbers of strain YXT and Pedobacter yonginense KCTC 22721T are QGNY00000000 and QGNZ00000000, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Li, J., Wang, J. et al. Pedobacter paludis sp. nov., isolated from wetland soil. Arch Microbiol 201, 349–355 (2019). https://doi.org/10.1007/s00203-018-1605-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1605-0