Abstract
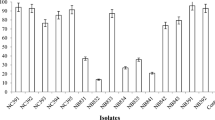
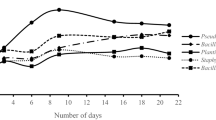
The aim of this work was to isolate and characterize bacteria from two crude oil-contaminated soils which were capable of producing biosurfactant and degrade oil. The contaminated soils were located in Tehran and Isfahan oil refineries, Iran. Various traits, such as hemolytic activity, oil resolving , oil spreading, and oil emulsifying capability were used to screen the isolates. Among 54 isolated isolates from two petroleum-contaminated soils, only 10 isolates were able to produce biosurfactant and degrade oil. These 10 isolates could produce glycolipid biosurfactants, which ranged from 1.93 to 3.7 g L−1. The results showed that four isolates (T4, T27, T30 and E1) could efficiently degrade crude oil in saline mineral broth, with the highest removal efficiency of 39% for T4. Gas chromatography analysis revealed that both lighter and heavier petroleum hydrocarbons were affected by microbial degradation. It was also demonstrated that branched alkanes as well as linear alkanes were degraded by the isolates. Based on 16S rDNA sequencing, all four isolates were identified as pseudomonas aeruginosa. Overall, this study shows that the biosurfactant production is a critical trait in screening of oil-degrading bacteria. Thus, the superior strains in this study are recommended for bioremediation programs, particularly in reclamation of crude oil-contaminated saline soils.





Similar content being viewed by others
References
Bento FM De, Oliveira Camargo FA, Okeke BC, Frankenberger WT (2005) Diversity of biosurfactant producing microorganisms isolated from soils contaminated with diesel oil. Microbiol Res 160:249–255
Betancur-Galvis LA, Alvarez-Bernal D, Ramos-Valdivia AC, Dendooven L (2006) Bioremediation of polycyclic aromatic hydrocarbon-contaminated saline–alkaline soils of the former Lake Texcoco. Chemosphere 62:1749–1760
Bordas F, Lafrance P, Villemur R (2005) Conditions for effective removal of pyrene from an artificially contaminated soil using Pseudomonas aeruginosa 57SJ rhamnolipids. Environ Pollut 138:69–76
Bouvet P, Ferraris L, Dauphin B, Popoff MR, Butel MJ, Aires J (2014) 16S rRNA gene sequencing, multilocus sequence analysis, and mass spectrometry identification of the proposed new species “Clostridium neonatale”. J Clin Microbiol 52(12):4129–4136
Čipinytė V, Grigiškis S, Šapokaitė D, Baškys E (2015) Production of biosurfactants by Arthrobacter sp. N3, a hydrocarbon degrading bacterium. Environment Technology Resources. Proceedings of the International Scientific and Practical Conference 1:68–75. doi:10.17770/etr2011vol1.888
Das N, Chandran P (2011) Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. doi:10.4061/2011/941810
Fountoulakis MS, Terzakis S, Kalogerakis N, Manios T (2009) Removal of polycyclic aromatic hydrocarbons and linear alkylbenzene sulfonates from domestic wastewater in pilot constructed wetlands and a gravel filter. Ecol Eng 35:1702–1709
George S, Jayachandran K (2013) Production and characterization of rhamnolipid biosurfactant from waste frying coconut oil using a novel Pseudomonas aeruginosa. J Appl Microbiol 114:373–383
Graj W, Lisiecki P, Szulc A, Chrzanowski Ł, Wojtera-Kwiczor J (2013) Bioaugmentation with petroleum-degrading consortia has a selective growth-promoting impact on crop plants germinated in diesel oil-contaminated soil. Water Air Soil Pollut 224:1–15
Hua F, Wang H (2012) Uptake modes of octadecane by Pseudomonas sp. DG17 and synthesis of biosurfactant. J Appl Microbiol 112:25–37
Kadali KK, Simons KL, Skuza PP, Moore RB, Ball AS (2012) A complementary approach to identifying and assessing the remediation potential of hydrocarbonoclastic bacteria. J Microbiol Methods 88:348–355
Lai CC, Huang YC, Wei YH, Chang JS (2009) Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J Hazard Mater 167:609–614
Li X, Zhao L, Adam M (2016) Biodegradation of marine crude oil pollution using a salt-tolerant bacterial consortium isolated from Bohai Bay. China. Mar Pollut Bull. 105:43–50
Margesin R, Schinner F (2001) Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl Microbiol Biotechnol 56:650–663
Minai-Tehrani D, Rohanifar P, Azami S (2015) Assessment of bioremediation of aliphatic, aromatic, resin, and asphaltene fractions of oil-sludge-contaminated soil. Int J Environ Sci Technol 12:1253–1260
Mukherjee AK, Bordoloi NK (2011) Bioremediation and reclamation of soil contaminated with petroleum oil hydrocarbons by exogenously seeded bacterial consortium: a pilot-scale study. Environ Sci Pollut Res 18:471–478
Onwurah INE, Ogugua VN, Onyike NB, Ochonogor AE, Otitoju OF (2007) Crude oil spills in the environment, effects and some innovative clean-up biotechnologies. Int J Environ Res 1:307–320
Rocha CA, Pedregosa AM, Laborda F (2011) Biosurfactant-mediated biodegradation of straight and methyl-branched alkanes by Pseudomonas aeruginosa ATCC 55925. AMB Express 1:1–10. doi:10.1186/2191-0855-1-9
Soleimani M, Afyuni M, Hajabbasi MA, Nourbakhsh F, Sabzalian MR, Christensen JH (2010) Phytoremediation of an aged petroleum contaminated soil using endophyte infected and non-infected grasses. Chemosphere 81:1084–1090
Soleimani M, Farhoudi M, Christensen JH (2013) Chemometric assessment of enhanced bioremediation of oil contaminated soils. J Hazard Mater 254:372–381
Szulc A, Ambrożewicz D, Sydow M, Ławniczak Ł, Piotrowska-Cyplik A, Marecik R, Chrzanowski Ł (2014) The influence of bioaugmentation and biosurfactant addition on bioremediation efficiency of diesel-oil contaminated soil: feasibility during field studies. J Environ Manag 132:121–128
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb Ecol 62:324–336
Ulrich AC, Guigard SE, Foght JM, Semple KM, Pooley K, Armstrong JE, Biggar KW (2009) Effect of salt on aerobic biodegradation of petroleum hydrocarbons in contaminated groundwater. Biodegradation 20:27–38
Varjani SJ, Rana DP, Jain AK, Bateja S, Upasani VN (2015) Synergistic ex situ biodegradation of crude oil by halotolerant bacterial consortium of indigenous strains isolated from on shore sites of Gujarat, India. Int Biodeterior Biodegrad 103:116–124
Wadekar SD, Kale SB, Lali AM, Bhowmick DN, Pratap AP (2012) Microbial synthesis of rhamnolipids by Pseudomonas aeruginosa (ATCC 10145) on waste frying oil as low cost carbon source. Prep Biochem Biotechnol 42:249–266
Whang LM, Liu PWG, Ma CC, Cheng SS (2008) Application of biosurfactants, rhamnolipid, and surfactin, for enhanced biodegradation of diesel-contaminated water and soil. J Hazard Mater 151:155–163
Wu T, Xie WJ, Yi YL, Li XB, Yang HJ, Wang J (2012) Surface activity of salt-tolerant Serratia spp. and crude oil biodegradation in saline soil. Plant Soil Environ 58:412–416
Yadav AK, Manna S, Pandiyan K, Singh A, Kumar M, Chakdar H, Srivastava AK (2016) Isolation and characterization of biosurfactant producing Bacillus sp. from diesel fuel-contaminated site. Microbiology 85:56–62
Yan-Zheng G, Wan-Ting L, Li-Zhong Z, Bao-Wei Z, Zheng QS (2007) Surfactant-enhanced phytoremediation of soils contaminated with hydrophobic organic contaminants: potential and assessment. Pedosphere 17:409–418
Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ (2012) Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 7(3):e33865. doi:10.1371/journal.pone.0033865
Zeng H, Zhang D, Lehne E, Zou F, Zuo JY (2012) Gas chromatograph applications in petroleum hydrocarbon fluids. INTECH Open Access Publisher, Rijeka, pp 363–388
Zhang X, Xu D, Zhu C, Lundaa T, Scherr KE (2012) Isolation and identification of biosurfactant producing and crude oil degrading Pseudomonas aeruginosa strains. Chem Eng Manag 209:138–146
Acknowledgements
The financial and facility support of this project by the Agricultural Biotechnology Research Institute of Iran (ABRII) is appreciated. The authors also appreciate Shahid Tondgooyan oil refinery for providing crude oil for this research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ebadi, A., Olamaee, M., Khoshkholgh Sima, N.A. et al. Isolation and Characterization of Biosurfactant Producing and Crude Oil Degrading Bacteria from Oil Contaminated Soils. Iran J Sci Technol Trans Sci 42, 1149–1156 (2018). https://doi.org/10.1007/s40995-017-0162-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-017-0162-8