Abstract
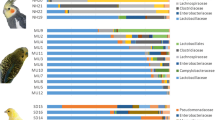
The primary goal of this study was to better understand the microbial composition and functional genetic diversity associated with turkey fecal communities. To achieve this, 16S rRNA gene and metagenomic clone libraries were sequenced from turkey fecal samples. The analysis of 382 16S rRNA gene sequences showed that the most abundant bacteria were closely related to Lactobacillales (47%), Bacillales (31%), and Clostridiales (11%). Actinomycetales, Enterobacteriales, and Bacteroidales sequences were also identified, but represented a smaller part of the community. The analysis of 379 metagenomic sequences showed that most clones were similar to bacterial protein sequences (58%). Bacteriophage (10%) and avian viruses (3%) sequences were also represented. Of all metagenomic clones potentially encoding for bacterial proteins, most were similar to low G+C Gram-positive bacterial proteins, particularly from Lactobacillales (50%), Bacillales (11%), and Clostridiales (8%). Bioinformatic analyses suggested the presence of genes encoding for membrane proteins, lipoproteins, hydrolases, and functional genes associated with the metabolism of nitrogen and sulfur containing compounds. The results from this study further confirmed the predominance of Firmicutes in the avian gut and highlight the value of coupling 16S rRNA gene and metagenomic sequencing data analysis to study the microbial composition of avian fecal microbial communities.
Similar content being viewed by others
References
Acinas, S.G., L.A. Marcelino, V. Klepac-Ceraj, and M.F. Polz. 2004. Diverge and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186, 2629–2635.
Althschul, S.F., F. Thomas, L. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L.N. Ornston, J. Weissenbach, P. Marlière, G.N. Cohen, and C. Médigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32, 5766–5779.
Birnbaum, D., L. Herwaldt, D.E. Low, M. Noble, M. Pfaller, R. Sherertz, and A. Chow. 1994. Efficacy of microbial identification system for epidemiologic typing of coagulase negative staphylococci. J. Clin. Microbiol. 32, 2113–2119.
Breitbart, M., P. Salamon, B. Andresen, J.M. Mahaffy, A.M. Segall, D. Mead, F. Azam, and F. Rohwer. 2003. Genomic analysis of uncultured marine viral communities. J. Bacteriol. 185, 6220–6223.
Buchan, A., E.L. Neidle, and M.A. Moran. 2001. Diversity of the ring-cleaving dioxygenase gene pcaH in a salt marsh bacterial community. Appl. Environ. Microbiol. 67, 5801–5809.
Case, R.J., Y. Boucher, I. Dahllöf, C. Holmström, W.F. Doolittle, and S. Kjelleberg. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73, 278–288.
Christiansen, N. and B.K. Ahring. 1996. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46, 442–448.
Cole, J.R., B. Chai, R.J. Farris, Q. Wang, A.S. Kulam-Syed-Mohideen, D.M. McGarrell, A.M. Bandela, E. Cardenas, G.M. Garrity, and J.M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35, D169–172.
Collins, M.D., R.A. Hutson, E. Falsen, E. Inganas, and M. Bisgaard. 2002. Streptococcus gallinaceus sp. nov., from chickens. Int. J. Syst. Evol. Microbiol. 52, 1161–1164.
Cotta, M.A., T.R. Whitehead, and R.L. Zeltwanger. 2003. Isolation, characterization and comparison of bacteria from swine feces and manure storage pits. Environ. Microbiol. 5, 737–745.
Crosby, L.D. and C.S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34, 790–794.
Daly, K., C.S. Stewart, H.J. Flint, and S.P. Shirazi-Beechey. 2001. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol. Ecol. 38, 141–151.
Eckburg, P.B., E.M. Bik, C.N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S.R. Gill, K.E. Nelson, and D.A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308, 1635–1638.
Edwards, E.J., N.R. McEwan, A.J. Travis, and R.J. Wallace. 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86, 263–281.
Facklam, R., M. Lovgren, P.L. Shewmaker, and G. Tyrrell. 2003. Phenotypic description and antimicrobial susceptibilities of Aerococcus sanguinicola isolates from human clinical samples. J. Clin. Microbiol. 41, 2587–2592.
Fairchild, A.S., J.L. Smith, U. Idris, J. Lu, S. Sanchez, L.B. Purvis, C. Hofacre, and M.D. Lee. 2005. Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71, 5865–5872.
Gill, S.R., M. Pop, R.T. Deboy, P.B. Eckburg, P.J. Turnbaugh, B.S. Samuel, J.I. Gordon, D.A. Relman, C.M. Fraser-Liggett, and K.E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359.
Gong, J., W. Si, R.J. Forster, R. Huang, H. Yu, Y. Yin, C. Yang, and Y. Han. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 59, 147–157.
Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319.
Iwabuchi, T. and S. Harayama. 1998. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J. Biol. Chem. 273, 8332–8336.
Janssen, P.H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728.
Joseph, S.W., J.R. Hayes, L.L. English, L.E. Carr, and D.D. Wagner. 2001. Implication of multiple antimicrobial-resistant enterococci associated with the poultry environment. Food Addit. Contam. 18, 1118–1123.
Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V.K. Sharma, T.P. Srivastava, T.D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D.S. Ehrlich, K. Itoh, T. Takagi, Y. Sakaki, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181.
Leser, T.D., J.Z. Amenuvor, T.K. Jensen, R.H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68, 673–690.
Liles, M.R., B.F. Manske, S.B. Bintrim, J. Handelsman, and R.M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69, 2684–2691.
Lu, J., U. Idris, B.G. Harmon, C. Hofacre, J.J. Maurer, and M.D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69, 6816–6824.
Lu, J., J.W. Santo Domingo, and O.C. Shanks. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41, 3561–3574.
Ludwig, W. and K.H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23, 556–562.
Marois, C., C. Savoye, M. Kobisch, and I. Kempf. 2002. A reverse transcription-PCR assay to detect viable Mycoplasma synoviae in poultry environmental samples. Vet. Microbiol. 89, 17–28.
Pearl, F.M.G., D. Lee, J.E. Bray, I. Sillitoe, A.E. Todd, A.P. Harrison, J.M. Thornton, and C.A. Orengo. 2000. Assigning genomic sequences to CATH. Nucleic Acids Res. 28, 277–282.
Rattray, F.P. and P.F. Fox. 1999. Aspects of enzymology and biochemical properties of Brevibacterium linens relevant to cheese ripening: a review. J. Dairy Sci. 82, 891–909.
Rondon, M.R., P.R. August, A.D. Bettermann, S.F. Brady, T.H. Grossman, M.R. Liles, K.A. Loiacono, B.A. Lynch, I.A. MacNeil, C. Minor, C.L. Tiong, M. Gilman, M.S. Osburne, J. Clardy, J. Handelsman, and R.M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547.
Rosato, A.E., B.S. Lee, and K.A. Nash. 2001. Inducible macrolide resistance in Corynebacterium jeikeium. Antimicrob. Agents Chemother. 45, 1982–1989.
Sanford, R.A., J.R. Cole, and J.M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic Myxobacterium. Appl. Environ. Microbiol. 68, 893–900.
Schmeisser, C., C. Stöckigt, C. Raasch, J. Wingender, K.N. Timmis, D.F. Wenderoth, H.C. Flemming, H. Liesegang, R.A. Schmitz, K-E. Jaeger, and W.R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69, 7298–7309.
Scupham, A.J. 2007a. Succession in the intestinal microbiota of preadolescent turkeys. FEMS Microbiol. Ecol. 60, 136–147.
Scupham, A.J. 2007b. Examination of the microbial ecology of the avian intestine in vivo using bromouridine. Environ. Microbiol. 9, 1801–1809.
Singleton, D.R., M.A. Furlong, S.L. Rathbun, and W.B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67, 4374–4376.
Suau, A., R. Bonnet, M. Sutren, J.J. Godon, G.R. Gibson, M.D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807.
Suzuki, M.T., L.T. Taylor, and E.F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614.
Tatusov, R.L., E.V. Koonin, and D.J. Lipman. 1997. A genomic perspective on protein families. Science 278, 631–637.
Tringe, S.G., C. Von Mering, A. Kobayashi, A.A. Salamov, K. Chen, H.W. Chang, M. Podar, J.M. Short, E.J. Mathur, J.C. Detter, P. Bork, P. Hugenholtz, and E.M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308, 554–557.
Zhong, Y., F. Chen, S.W. Wilhelm, L. Poorvin, and R.E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68, 1576–1584.
Zhu, X.Y., T. Zhong, Y. Panya, and R.D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68, 124–137.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, J., Domingo, J.S. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J Microbiol. 46, 469–477 (2008). https://doi.org/10.1007/s12275-008-0117-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-008-0117-z