Abstract
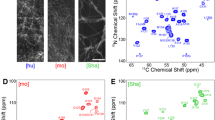
Sup35pNM represents the N-terminal and middle (M) domains of the yeast Saccharomyces cerevisiae prion Sup35p. This fragment is commonly used for structural and functional studies of Sup35p. We here present a solid-state NMR study of fibrils formed by this fragment and show that sequential assignments can be obtained for the rigid and well-ordered parts of the protein using 3D spectroscopy. We describe in detail the sequential assignment of the 22 residues yielding strong, narrow signals with chemical shifts that correspond mostly to β-sheet secondary-structured amino acids that form the fibril core.






Similar content being viewed by others
References
Böckmann A, Gardiennet C, Verel R, Hunkeler A, Loquet A, Pintacuda G, Emsley L, Meier BH, Lesage A (2009) Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR 45(3):319–327
Chien P, Yonekura K, Weissman J (2005) Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121:49–62
Cox BS (1965) Psi, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20(4):505–521
DePace AH, Santoso A, Hillner P, Weissman JS (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93(7):1241–1252
Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, Henrick K, Gilliland G, Berman H, Thornton J, Nilges M, Markley J, Laue E (2002) The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol 9(6):416–418
Foo CK, Ohhashi Y, Kelly MJS, Tanaka M, Weissman JS (2011) Radically different amyloid conformations dictate the seeding specificity of a chimeric Sup35 prion. J Mol Biol 408(1):1–8
Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89(5):811–819
Habenstein B, Wasmer C, Bousset L, Sourigues Y, Schütz A, Loquet A, Meier BH, Melki R, Böckmann A (2011) Extensive de novo solid-state NMR assignments of the 33 kDa C-terminal domain of the Ure2 prion. J Biomol NMR 51(3):235–243
Halfmann R, Alberti S, Krishnan R, Lyle N, O’Donnell CW, Donnell CW, King OD, Berger B, Pappu RV, Lindquist S (2011) Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell 43(1):72–84
Krishnan R, Lindquist S (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435(7043):765–772
Krzewska J, Melki R (2006) Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J 25(4):822–833
Krzewska J, Tanaka M, Burston SG, Melki R (2007) Biochemical and functional analysis of the assembly of full-length Sup35p and its prion-forming domain. J Biol Chem 282(3):1679–1686
Luca S, Filippov D, van Boom J, Oschkinat H, de Groot H, Baldus M (2001) Secondary chemical shifts in immobilized peptides and proteins: a qualitative basis for structure refinement under magic angle spinning. J Biomol NMR 20(4):325–331
Luckgei N, Schütz AK, Bousset L, Habenstein B, Sourigues Y, Gardiennet C, Meier BH, Melki R, Böckmann A (2013) The conformation of the prion domain of Sup35p in isolation and in the full-length protein is different (under review)
Schuetz A, Wasmer C, Habenstein B, Verel R, Greenwald J, Riek R, Böckmann A, Meier BH (2010) Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s (1-227). ChemBioChem 11(11):1543–1551
Schütz AK, Habenstein B, Luckgei N, Bousset L, Sourigues Y, Nielsen AB, Melki R, Böckmann A, Meier BH (2013) Solid-state NMR sequential assignments of the amyloid core of full-length Sup35p (submitted)
Stevens TJ, Fogh RH, Boucher W, Higman VA, Eisenmenger F, Bardiaux B, van Rossum B-J, Oschkinat H, Laue ED (2011) A software framework for analysing solid-state MAS NMR data. J Biomol NMR 51(4):437–447
Toyama B, Kelly M, Gross J, Weissman J (2007) The structural basis of yeast prion strain variants. Nature 449(7159):233–237
Verges KJ, Smith MH, Toyama BH, Weissman JS (2011) Strain conformation, primary structure and the propagation of the yeast prion [PSI+]. Nat Struct Mol Biol 18(4):493–499
Vranken W, Boucher W, Stevens T, Fogh R, Pajon A, Llinas P, Ulrich E, Markley J, Ionides J, Laue E (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59(4):687–696
Wang Y, Jardetzky O (2002) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci 11(4):852–861
Wickner RB, Masison DC, Edskes HK (1995) [PSI] and [URE3] as yeast prions. Yeast 11(16):1671–1685
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4(2):171–180
Acknowledgments
We thank Dr. Christian Wasmer for help with recording the spectra. This work was supported by the Agence Nationale de la Recherche (ANR-12-BS08-0013-01), the ETH Zurich, the Swiss National Science Foundation (Grant 200020_124611) and the Centre National de la Recherche Scientifique. We also acknowledge support from the European Commission under the Seventh Framework Programme (FP7), contract Bio-NMR 261863.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Nina Luckgei, Anne Schütz, Birgit Habenstein, and Luc Bousset have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Luckgei, N., Schütz, A.K., Habenstein, B. et al. Solid-state NMR sequential assignments of the amyloid core of Sup35pNM. Biomol NMR Assign 8, 365–370 (2014). https://doi.org/10.1007/s12104-013-9518-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-013-9518-y