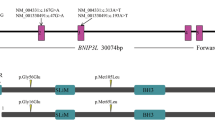
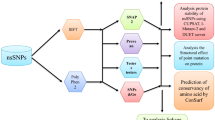
Abstract
Catechol-O-methyltransferase (COMT) enzyme performs transfer of methyl group to endogenous and exogenous catechol substrates. The COMT enzyme draws interest because of its association with psychiatric, neurological and cardiovascular diseases, and several cancers. Moreover, many prescribed drugs, supplements, and their metabolites are used as substrates of COMT enzyme. The human COMT gene has 226 nonsynonymous single nucleotide polymorphisms (nsSNPs) according to public databases. Uncovering of the molecular impacts of nsSNPs on COMT enzyme function and structure may provide standpoint on how COMT nsSNPs affect enzyme activity and contribute to disease development. Therefore, we aimed in this study to predict possible structural and functional damaging effects of all knowns nsSNPs in COMT gene by applying various bioinformatics tools. Two hundred and twenty-six nsSNPs were obtained from Ensembl, HGMD, ClinVar, and dbSNP databases. Twenty-eight nsSNPs were found to be high-risk changes for protein structure. Some of them were detected in extremely conserved sequences have functional and structural properties. Besides, high-risk nsSNPs were also uncovered to change properties of native COMT protein. Our findings demonstrated the significance of COMT high-risk nsSNPs on protein structure and function. We expect that our results will be helpful in future studies concerning experimental evaluation of the COMT gene polymorphisms and/or the association between COMT polymorphisms and disease development.

Similar content being viewed by others
References
Mao, C., Wang, X.-W., Qiu, L.-X., Liao, R.-Y., Ding, H., & Chen, Q. (2010). Lack of association between catechol-O-methyltransferase Val108/158Met polymorphism and breast cancer risk: a meta-analysis of 25,627 cases and 34,222 controls. Breast Cancer Research and Treatment, 121(3), 719–725.
Sak, K. (2017). The Val158Met polymorphism in COMT gene and cancer risk: role of endogenous and exogenous catechols. Drug Metabolism Reviews, 49(1), 56–83.
Hall, K. T., Loscalzo, J., & Kaptchuk, T. J. (2019). Systems pharmacogenomics—gene, disease, drug and placebo interactions: a case study in COMT. Pharmacogenomics, 20(7), 529–551.
Brahe, C., Bannetta, P., Meera, K. P., Arwert, F., & Serra, A. (1986). Assignment of the catechol-O-methyltransferase gene to human chromosome 22 in somatic cell hybrids. Human Genetics, 74(3), 230–4.
Winqvist, R., Lundstrom, K., Salminen, M., Laatikainen, M., & Ulmanen, I. (1991). Mapping of human catechol –O methyltransferase gene to 22q11.2 and detection of a frequent RFLP with BglI. Cytogenetics and Cell Genetics58(3–4), 2051.
Tenhunen, J., Salminen, M., Lundström, K., Kiviluoto, T., Savolainen, R., & Ulmanen, I. (1994). Genomic organization of the human catechol-O-methyltransferase gene and its expression from two distinct promoters. European Journal of Biochemistry, 223(3), 1049–1059.
Lundström, K., Tenhunen, J., Tilgmann, C., Karhunen, T., Panula, P., & Ulmanen, I. (1995). Cloning, expression and structure of catechol-O-methyltransferase. Biochimica et Biophysica Acta, 1251(1), 1–10.
Lotta, T., Vidgren, J., Tilgmann, C., Ulmanen, I., Melén, K., & Julkunen, I., et al. (1995). Kinetics of human soluble and membrane-bound catechol-O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry, 34(13), 4202–4210.
Matsumoto, M., Weickert, C. S., Beltaifa, S., Kolachana, B., Chen, J., & Hyde, T. M., et al. (2003). Catechol O-methyltransferase (COMT) mRNA expression in the dorsolateral prefrontal cortex of patients with schizophrenia. Neuropsychopharmacology, 28(8), 1521–1530.
Bastos, P., Gomes, T., & Ribeiro, L. (2017). Catechol-O-Methyltransferase (COMT): An Update on Its Role in Cancer, Neurological and Cardiovascular Diseases. In B. Nilius, P. de Tombe, T. Gudermann, R. Jahn, R. Lill, & O. H. Petersen (Eds.), Reviews of Physiology, Biochemistry and Pharmacology, Springer International Publishing, Cham, Switzerland, Vol. 173 (pp. 1–39).
Robert, F., & Pelletier, J. (2018). Exploring the impact of single-nucleotide polymorphisms on translation. Frontiers in Genetics, 9, 507.
Cunningham, F., Achuthan, P., Akanni, W., Allen, J., Amode, M. R., & Armean, I. M., et al. (2019). Ensembl 2019. Nucleic Acids Research, 47(D1), D745–D751.
Stenson, P. D., Mort, M., Ball, E. V., Shaw, K., Phillips, A. D., & Cooper, D. N. (2014). The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Human Genetics, 133(1), 1–9.
Landrum, M. J., Lee, J. M., Riley, G. R., Jang, W., Rubinstein, W. S., Church, D. M., & Maglott, D. R. (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research, 42, D980–D985.
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., & Sirotkin, K. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Research, 29(1), 308–311.
The UniProt Consortium. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Research, 45(Database issue), D158–D169.
Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C., Chen, L., & Costanzo, L. D., et al. (2019). Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Research, 47(D1), D520–D528.
Sim, N.-L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., & Ng, P. C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Research, 40(W1), W452–W457.
Choi, Y., & Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics, 31(16), 2745–2747.
Reva, B., Antipin, Y., & Sander, C. (2011). Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Research, 39(17), e118.
Mi, H., Huang, X., Muruganujan, A., Tang, H., Mills, C., & Kang, D., et al. (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research, 45(Database issue), D183–D189.
Hecht, M., Bromberg, Y., & Rost, B. (2015). Better prediction of functional effects for sequence variants. BMC Genomics, 16(Suppl 8), S1.
Yates, C. M., Filippis, I., Kelley, L. A., & Sternberg, M. J. E. (2014). SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. Journal of Molecular Biology, 426(14), 2692–2701.
González-Pérez, A., & López-Bigas, N. (2011). Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. American Journal of Human Genetics, 88(4), 440–449.
Capriotti, E., Altman, R. B., & Bromberg, Y. (2013). Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics, 14(Suppl 3), S2.
Bendl, J., Stourac, J., Salanda, O., Pavelka, A., Wieben, E. D., & Zendulka, J., et al. (2014). PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Computational Biology, 10(1), e1003440.
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., & Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7(4), 248–249.
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., & Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research, 44(W1), W344–W350.
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R., & Wu, C. H. (2007). UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics, 23(10), 1282–1288.
Capriotti, E., Fariselli, P., & Casadio, R. (2005). I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Research, 33(Web Server issue), W306–W310.
Pires, D. E. V., Ascher, D. B., & Blundell, T. L. (2014). DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Research, 42(W1), W314–W319.
Klausen, M. S., Jespersen, M. C., Nielsen, H., Jensen, K. K., Jurtz, V. I., & Sønderby, C. K., et al. (2019). NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins: Structure, Function, and Bioinformatics, 87(6), 520–527.
Venselaar, H., te Beek, T. A., Kuipers, R. K., Hekkelman, M. L., & Vriend, G. (2010). Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics, 11(1), 548.
Pejaver, V., Urresti, J., Lugo-Martinez, J., Pagel, K. A., Lin, G. N., & Nam, H.-J., et al. (2017). MutPred2: Inferring the molecular and phenotypic impact of amino acid variants. BioRxiv 134981, https://doi.org/10.1101/134981.
Kabsch, W., & Sander, C. (1983). Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22(12), 2577–2637.
Klose, D. P., Wallace, B. A., & Janes, R. W. (2010). 2Struc: the secondary structure server. Bioinformatics, 26(20), 2624–2625.
Kaplan, W., & Littlejohn, T. G. (2001). Swiss-PDB Viewer (Deep View). Briefings in Bioinformatics, 2(2), 195–197.
Ng, P. C., & Henikoff, S. (2006). Predicting the effects of amino acid substitutions on protein function. Annual Review of Genomics and Human Genetics, 7(1), 61–80.
Singh, R. K., & Mahalingam, K. (2017). In silico approach to identify non-synonymous SNPs in human obesity related gene, MC3R (melanocortin-3-receptor). Computational Biology and Chemistry, 67, 122–130.
Singh, H. B., Deka, D., Das, D., & Borbora, D. (2017). Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in the human Quinone Oxidoreductase 1 (NQO1). Meta Gene, 11, 127–135.
Pires, A. S., Porto, W. F., Franco, O. L., & Alencar, S. A. (2017). In silico analyses of deleterious missense SNPs of human apolipoprotein E3. Scientific Reports, 7(1), 2509.
Yilmaz, A. (2015). Bioinformatic analysis of GJB2 gene missense mutations. Cell Biochemistry and Biophysics, 71(3), 1623–1642.
Chen, J., Lipska, B. K., Halim, N., Ma, Q. D., Matsumoto, M., & Melhem, S., et al. (2004). Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. American Journal of Human Genetics, 75(5), 807–821.
Rutherford, K. J., Alphandery, E., McMillan, A., Dagget, V., & Parson, W. W. (2008). The V108M mutation decreases the structural stability of catechol-O-methyltransferase. Biochimica Et Biophysica Acta, 1784(7–8), 1098–1105.
Doyle, A. E., Goodman, J. E., Silber, P. M., & Yager, J. D. (2004). Catechol-O-methyltransferase low activity genotype (COMTLL) is associated with low levels of COMT protein in human hepatocytes. Cancer Letters, 214(2), 189–195.
Shield, A. J., Thomae, B. A., Eckloff, B. W., Wieben, E. D., & Weinshilboum, R. M. (2004). Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Molecular Psychiatry, 9(2), 151–160.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yilmaz, A., Çetin, İ. In Silico Prediction of the Effects of Nonsynonymous Single Nucleotide Polymorphisms in the Human Catechol-O-Methyltransferase (COMT) Gene. Cell Biochem Biophys 78, 227–239 (2020). https://doi.org/10.1007/s12013-020-00905-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-020-00905-6