Abstract
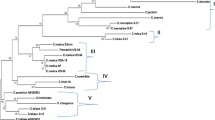
We sequenced the rbcL genes of 64 accessions from 24 genera of Citrus relatives and analyzed them by neighbor-joining and maximum parsimony methods. Both trees supported Swingle and Reece’s (1967) treatment of the subfamily Aurantioideae as monophyletic. However, the trees did not support Swingle and Reece’s treatment of tribes and subtribes. The subgenera Citrus and Papeda were not clustered clearly. The analysis associated the Fortunella group with mandarin, Poncirus with Citrus ichangensis, Severinia buxifolia with Atalantia ceylanica, Microcitrus with Eremocitrus and Citrus micrantha, and Hesperethusa crenulata with Citropsis. Furthermore, Atalantia species showed polytomy. The classification of Swingle and Reece should be reviewed.


Similar content being viewed by others
References
Aradhya MK, Potter D, Gao F, Simon CJ (2007) Molecular phylogeny of Juglans (Juglandaceae): a biogeographic perspective. Tree Genetics & Genomes 3:363–378
Araújo EF, Queiroz LP, Machado MA (2003) What is Citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae). Org Divers Evol 3:55–62
Asadi Abkenar A, Isshiki S, Tashiro Y (2004) Phylogenetic relationships in the “true citrus fruit trees” revealed by PCR-RFLP analysis of cpDNA. Sci Hort 102:233–242
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Barrett HC, Rhodes AM (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1:105–136
Bayer RJ, Mabberley DJ, Morton C, Miller CH, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S (2009) A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot 96:668–685
Chase MW, Morton CM, Kallunki JA (1999) Phylogenetic relationships of Rutaceae: a cladistic analysis of the subfamilies using evidence from rbcL and atpB sequence variation. Am J Bot 86:1191–1199
Cheng Y, Vicente MR, Meng H, Guo W, Tao N, Deng X (2005) A set of primers for analyzing chloroplast DNA diversity in Citrus and related genera. Tree Physiol 25:661–672
Deng Z, La Malfa S, Xie Y, Xiong X, Gentile A (2007) Identification and evolution of chloroplast uni- and trinucleotide sequence repeats in citrus. Sci Hort 111:186–192
Eck RV, Dayhoff MO (1966) Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152:363–366
Fang DQ, Roose ML (1997) Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet 95:408–417
Federici CT, Fang DQ, Scora RW, Roose ML (1998) Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96:812–822
Fitch WN (1971) The nonidentity of invariable positions in the cytochromes c of different species. Biochem Genet 5:231–241
Gielly L, Taberlet P (1994) The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. Mol Biol Evol 11:769–777
Groppo MG, Pirani JR, Salatino MLF, Blanco SR, Kallunki JA (2008) Phylogeny of Rutaceae based on two noncoding regions from cpDNA. Am J Bot 95:985–1005
Guerra M, dos Santos KGB, e Silva AEB, Ehrendorfer F (2000) Heterochromatin banding patterns in Rutaceae–Aurantioideae. A case of parallel chromosomal evolution. Am J Bot 87:735–747
Hasebe M, Omori T, Kanazawa M, Sano T, Kato M, Iwatsuki K (1994) rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proc Natl Acad Sci USA 91:5730–5734
Herrero R, Asins MJ, Pina JA, Carbonell EA, Navarro L (1996) Genetic diversity in the orange subfamily Aurantioideae. II. Genetic relationships among genera and species. Theor Appl Genet 93:1327–1334
Jung YH, Kwon HM, Kang SH, Kang JH, Kim SC (2005) Investigation of the phylogenetic relationships within the genus Citrus (Rutaceae) and related species in Korea using plastid trnL-trnF sequences. Sci Hort 104:179–188
Kellogg E, Juliano ND (1997) The structure and function of RuBisCo and their implications for systematic studies. Am J Bot 84:413–428
Mabberley DJ (1998) Australian Citreae with notes on other Aurantioideae (Rutaceae). Telopea 7:333–344
Mabberley DJ (2004) Citrus (Rutaceae): a review of recent advances in etymology, systematics and medical applications. Blumea 49:481–498
Moore WS (1995) Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49:718–726
Morton CM, Grant M, Blackmore S (2003) Phylogenetic relationships of the Aurantioideae inferred from chloroplast DNA sequence data. Am J Bot 90:1463–1469
Murray GC, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acid Res 8:4321–4325
Nicolosi E, Deng ZN, Gentile A, La Malfa Continella SG, Tribulato E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Pang XM, Hu CG, Deng XX (2007) Phylogenic relationships within Citrus and its related genera as inferred from AFLP markers. Genet Res Crop Evol 54:429–436
Platt AR, Woodhall RW, George AL (2007) Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques 43:58–62
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Samuel RF, Ehrendorfer MW, Chase GH (2001) Phylogenetic analyses of Aurantioideae (Rutaceae) based on non-coding plastid DNA sequences and phytochemical features. Plant Biol 3:77–87
Silva MFDGF, Gottlieb OR, Ehrendorfer F (1988) Chemosystematics of Rutaceae: suggestions for a more natural taxonomy and evolutionary interpretation of the family. Pl Syst Evol 161:97–124
Stace HM, Armstrong JA, James SH (1993) Cytoevolutionary patterns in Rutaceae. Pl Syst Evol 187:1–28
Suyama Y, Yoshimaru H, Tsumura Y (2000) Molecular phylogenetic position of Japanese Abies (Pinaceae) based on chloroplast sequences. Mol Phylogenet Evol 16:271–277
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD (eds) The citrus industry, vol 1. University of California, Berkeley, pp 190–430
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanaka T (1977) Fundamental discussion of citrus classification. Studia Citrologia 14:1–5
Yamamoto M, Kobayashi S (1996) Polymorphism of chloroplast DNA in citrus. J Jpn Soc Hort Sci 65:291–296
Yamamoto M, Kobayashi S, Nakamura Y, Yamada Y (1993) Phylogenic relationships of citrus revealed by RFLP analysis of mitochondrial and chloroplast DNA. Jpn J Breed 43:355–365
Yamamoto M, Asadi Abkenar A, Matsumoto R, Nesumi H, Yoshida T, Kuniga T, Kubo T, Tominaga S (2007) CMA banding patterns of chromosomes in major Citrus species. J Jpn Soc Hort Sci 76:36–40
Yamamoto M, Asadi Abkenar A, Matsumoto R, Kubo T, Tominaga S (2008) CMA staining analysis of chromosomes in several species of Aurantioideae. Genet Resour Crop Evol 55:1167–1173
Zhang D-X, Hartley TG, Mabberley DJ (2008) Rutaceae. http://flora.huh.harvard.edu.china/mss/volume11/Rutaceae.pdf
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Gmitter
Rights and permissions
About this article
Cite this article
Penjor, T., Anai, T., Nagano, Y. et al. Phylogenetic relationships of Citrus and its relatives based on rbcL gene sequences. Tree Genetics & Genomes 6, 931–939 (2010). https://doi.org/10.1007/s11295-010-0302-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0302-1