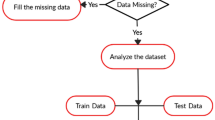
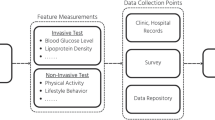
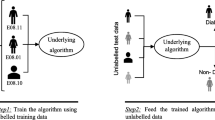
Abstract
Continuous ambulatory peritoneal dialysis (CAPD) is a treatment used by patients in the end-stage of chronic kidney diseases. Those patients need to be monitored using blood tests and those tests can present some patterns or correlations. It could be meaningful to apply data mining (DM) to the data collected from those tests. To discover patterns from meaningless data, it becomes crucial to use DM techniques. DM is an emerging field that is currently being used in machine learning to train machines to later aid health professionals in their decision-making process. The classification process can found patterns useful to understand the patients’ health development and to medically act according to such results. Thus, this study focuses on testing a set of DM algorithms that may help in classifying the values of serum creatinine in patients undergoing CAPD procedures. Therefore, it is intended to classify the values of serum creatinine according to assigned quartiles. The better results obtained were highly satisfactory, reaching accuracy rate values of approximately 95%, and low relative absolute error values.


Similar content being viewed by others
References
Rodrigues, M., Peixoto, H., Esteves, M., Machado, J., & Abelha, A. (2017). Understanding stroke in dialysis and chronic kidney disease. Procedia Computer Science, 113, 591–596.
Venkatapathy, R., Govindarajan, V., Oza, N., Parameswaran, S., Pennagaram Dhanasekaran, B., & Prashad, K. V. (2014). Salivary creatinine estimation as an alternative to serum creatinine in chronic kidney disease patients. International Journal of Nephrology, 2014, 1–6.
Guyton, A. C., & Hall, J. E. (2006). Guyton and hall textbook of medical physiology. Amsterdam: Elsevier.
Fink, J. C., Burdick, R. A., Kurth, S. J., Blahut, S. A., Armistead, N. C., Turner, M. S., et al. (1999). Significance of serum creatinine values in new end-stage renal disease patients. The American Journal of Kidney Diseases, 34, 694–701.
Davis, C. P., & Shield Jr., W. C. (2018). Creatinine (low, high, blood test results explained). https://www.medicinenet.com/creatinine_blood_test/article.htm#what_is_creatinine. Accessed 21 Jan 2019.
Mildred Lam, M. (2018). Kidney failure—Understanding end stage renal disease (ESRD). http://www.netwellness.org/healthtopics/kidney/kidney2.cfm. Accessed 21 Jan 2019.
Peake, M., & Whiting, M. (2006). Measurement of serum creatinine—Current status and future goals. The Clinical Biochemist Reviews, 27, 173–184.
Oliveira, P., Portela, F., Santos, M. F., Machado, J., Abelha, A., Silva, Á., & Rua, F. (2016). Optimization techniques to detect early ventilation extubation in intensive care units. In Advances in Intelligent Systems and Computing (AISC) (pp. 599–608). Cham: Springer.
Domingos, P. (2012). A few useful things to know about machine learning. Communications of the ACM, 55, 78–87.
Abernethy, M. (2010). Data mining with WEKA, Part 2: Classification and clustering. https://www.ibm.com/developerworks/library/os-weka2/. Accessed 21 Jan 2019.
Veloso, R., Portela, F., Santos, M. F., Machado, J., da Silva Abelha, A., Rua, F., et al. (2017). Categorize readmitted patients in intensive medicine by means of clustering data mining. International Journal of E-Health and Medical Communications, 8, 22–37.
Naik, A., & Samant, L. (2016). Correlation review of classification algorithm using data mining tool: WEKA. Procedia Computer Science, 85, 662–668.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., & Witten, I. H. (2009). The WEKA data mining software. ACM SIGKDD Explorations Newsletter, 11, 10–18.
Blobel, B. (2002). Analysis, design and implementation of secure and interoperable distributed health information systems. Studies in Health Technology and Informatics, 89, 1–352.
Portela, F., Santos, M. F., Machado, J., Abelha, A., Rua, F., & Silva, Á. (2015). Real-time decision support using data mining to predict blood pressure critical events in intensive medicine patients. In Lecture Notes in Computer Science (LNCS) (pp. 77–90). New York: Springer.
Portela, F., Filipe Santos, M., Silva, A., Rua, F., Abelha, A., & Machado, J. (2014). Preventing patient cardiac arrhythmias by using data mining techniques. In 2014 IEEE Conference on Biomedical Engineering and Sciences (IECBES). IEEE (2014) (pp. 165–170).
Pereira, S., Portela, F., Santos, M., Machado, J., & Abelha, A. (2016). Predicting pre-triage waiting time in a maternity emergency room through data mining. In Lecture Notes in Computer Science (LNCS)—Smart Health. New York: Springer.
Oliveira, S., Portela, F., Santos, M. F., Machado, J., & Abelha, A. (2014). Predictive models for hospital bed management using data mining techniques. In Advances in Intelligent Systems and Computing (AISC) (pp. 407–416). New York: Springer.
Aqlan, F., Markle, R., & Shamsan, A. (2017). Data mining for chronic kidney disease prediction. In Industrial and Systems Engineering Research Conference (ISERC).
Sharma, S., Sharma, V., & Sharma, A. (2016). Performance based evaluation of various machine learning classification techniques for chronic kidney disease diagnosis. International Journal of Modern Computer Scienc, 4, 11–16.
Bala, S., & Kumar, K. (2014). A literature review on kidney disease prediction using data mining classification technique. International Journal of Computer Science and Mobile Computing, 37, 960–967.
Baker, K., Dunwoodie, E., Jones, R. G., Newsham, A., Johnson, O., Price, C. P., et al. (2017). Process mining routinely collected electronic health records to define real-life clinical pathways during chemotherapy. International Journal of Medical Informatics, 103, 32–41.
Chawla, N. V. (2005). Data mining and knowledge discovery handbook. New York: Springer.
Vijayarani, S., & Muthulakshmi, M. (2013). Comparative analysis of bayes and lazy classification algorithms. International Journal of Advanced Research in Computer and Communication Engineering, 2, 3118–3124.
Tejera Hernández, D. C. (2015). An experimental study of K* algorithm. International Journal of Information Engineering and Electronic Business, 7, 14–19.
Horning, N. (2010). Random forests: An algorithm for image classification and generation of continuous fields data sets. In The International Conference on GeoInformatics for Spatial-Infrastructure Development in Earth & Allied Sciences 2010 (pp. 1–6).
Devasena, L. (2014). Comparative analysis of random forest, REP tree and J48 classifiers for credit risk prediction. In IJCA Proceedings on International Conference on Communication, Computing and Information Technology (pp. 30–36).
Breiman, L., & Cutler, A. (2018). Random forests—Classification description. https://www.stat.berkeley.edu/~breiman/RandomForests/cc_home.htm. Accessed 21 Jan 2019.
Kalmegh, S. (2015). Analysis of WEKA data mining algorithm REPTree, SimpleCart and RandomTree for classification of indian news. International Journal of Innovative Science Engineering and Technology, 2, 438–446.
Bengio, Y., & Grandvalet, Y. (2004). No unbiased estimator of the variance of K-fold cross-validation. Journal of Machine Learning Research, 5, 1089–1105.
Acknowledgements
This work has been supported by Compete POCI-01-0145—FEDER-007043 and FCT—Fundação para a Ciência e Tecnologia within the Project Scope UID/CEC/00319/2013.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brito, C., Esteves, M., Peixoto, H. et al. A data mining approach to classify serum creatinine values in patients undergoing continuous ambulatory peritoneal dialysis. Wireless Netw 28, 1269–1277 (2022). https://doi.org/10.1007/s11276-018-01905-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11276-018-01905-4