Abstract
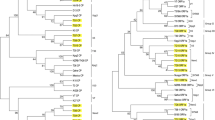
Citrus tristeza virus (CTV) isolates representing all the citrus-growing geographical zones of India were analyzed for nucleotide sequence of the 5′ORF1a fragments of the partial LProI domain and for the coat protein (CP) gene. The nucleotide sequences were compared with previously reported Indian and CTV genotypes from GenBank. The Indian isolates had 80–99 % sequence identity for the 5′ORF1a and 89–99 % identity for the CP genes. In phylogenetic tree analysis, all the Indian and previously reported isolates segregated into eight clades or groups for the 5′ORF1a region. Indian CTV isolates were clustered in all the clades, four of which, D13, K5, BAN-1, and B165, consisted of only Indian isolates. Phylogenetic tree analysis of the CP genes resulted in seven clades. Indian CTV isolates clustered in six of them, and clades I and VI consisted of only Indian isolates. In the phylogenetic tree the Indian CTV isolates clustered in different groups regardless their geographical origin. Diversities in CTV isolates within individual citrus farms were highlighted. Because incongruent phylogenetic relationships were observed for both of the genomic regions, 5′ORF1a and CP gene, recombination analysis was performed using program RDP3. This analysis detected potential recombination events among the CTV isolates which involved exchange of sequences between divergent CTV variants. The SplitsTree analysis showed evidence of phylogenetic conflicts in evolutionary relationships among CTV isolates.


Similar content being viewed by others
References
M. Bar-Joseph, W.O. Dawson, Encycl. Virol. 1, 520–525 (2008)
M. Bar-Joseph, R.F. Lee, in Description of plant viruses (Association of Applied Biology, Wellesbourne, 1989)
M. Bar-Joseph, R. Marcus, R.F. Lee, Annu. Rev. Phytopathol. 27, 291–316 (2002)
V. Karasev, V.P. Boyko, S. Gowda, O.V. Nikolaeva, M.E. Hilf, E.V. Koonin, K. Cline, D.J. Gumpf, R.F. Lee, S.M. Garnsey, D.J. Lewandowski, W.O. Dawson, Virology 208, 511–520 (1995)
R.F. Lee, M. Bar-Joseph, in Compendium of citrus diseases, 2nd edn., ed. by L.W. Timmer, S.M. Garnsey, J.H. Graham (APS, St. Paul, 2000), p. 61
P. Moreno, S. Ambros, M.R. Albiach-Marti, J. Guerri, L. Pena, Mol. Plant Pathol. 9, 251–268 (2008)
Anonymous, Horticulture database (National Horticulture Board, Gurgaon, 2009)
N. K. Chakraborty, Y.S. Ahlawat, A. Varma, K. J. Chandra, S. Ramapandu, S. P. Kapur, in Proceeding of the 12th Conference of the International Organization of Citrus Virologists, India (IOCV, Riverside, 1992), p. 108
Y.S. Ahlawat, Indian J. Agric. Sci. 67, 51–57 (1997)
K.K. Biswas, Indian J. Virol. 19, 26–31 (2008)
D.K. Ghosh, B. Aglave, V.K. Baranwal, Curr. Sci. 94, 1314–1318 (2008)
L. Rubio, M.A. Ayllon, P. Kong, A. Fernandez, M. Polek, J. Guerri, P. Moreno, B.W. Falk, J. Virol. 75, 8054–8062 (2001)
M.E. Hilf, V.A. Mavrodieva, Phytopathology 95, 909–917 (2005)
A. Roy, K.L. Manjunath, R.H. Brlansky, Virus Res. 113, 132–142 (2005)
S. Martin, A. Sambade, L. Rubio, M.C. Vives, P. Moya, J. Guerri, S.F. Elena, P. Moreno, J. Gen. Virol. 90, 1527–1538 (2009)
K.K. Biswas, Arch. Virol. 155, 959–963 (2010)
M.J. Melzer, W.B. Sether, D.M. Borth, S. Ferreira, D. Gonsalves, J.S. Hu, Virus Genes 40, 111–118 (2010)
A. Roy, R.H. Brlansky, Virus Res. 151, 118–130 (2010)
K.K. Biswas, A. Tarafdar, S.K. Sharma, Arch. Virol. (2011). doi:10.1007/s00705-011-1165-y
C. Lopez, M.A. Ayllon, J. Navas-Castillo, J. Guerri, P. Moreno, R. Flores, Phytopathology 88, 685–691 (1998)
M.C. Vives, L. Rubio, C. Lopez, J. Navas-Castillo, M.R. Albiach-Marti, W.O. Dawson, R. Flores, P. Moreno, J. Gen. Virol. 80, 811–816 (1999)
F. Garcia-Arenal, A. Fraile, J.M. Malpica, Annu. Rev. Phytopathol. 39, 157–186 (2001)
M.C. Vives, L. Rubio, A. Sambade, T.E. Mirkov, P. Moreno, J. Guerri, J Virol. 331, 297–307 (2005)
A. Roy, P. Ramachandran, R.H. Brlansky, Arch. Virol. 148, 207–722 (2003)
S.K. Sharma, A. Tarafdar, D. Khatun, S. Kumari, K.K. Biswas, J. Plant Biochem. Biotechnol. (2011). doi:10.1007/s13562-011-0071-4
J.D. Thompson, T.J. Gibson, F. Plewniak, F. Jeanmougin, D.J. Higgins, Nucleic Acids Res. 24, 4876–4882 (1997)
K. B Nicholas, H.B. Jr. Nicholas, www.psc.edu/biomed/genedoc
K. Tamura, J. Dudley, M. Nei, S. Kumar, Mol. Biol. Evol. 24, 1596–1599 (2007)
F. Tajima, Genetics 123, 585–595 (1989)
D.P. Martin, P. Lemey, M. Lott, V. Moulton, D. Posada, Bioinformatics 26, 2462–2463 (2010)
D.H. Huson, Bioinformatics 14, 68–73 (1998)
S.K. Thind, P.K. Arora, N. Kaur, J.K. Arora, Indian J. Virol. 16, 15–16 (2005)
K. L. Manjunath, R. F. Lee, C. L. Niblett, in Proceeding of the 14th Conference of the International Organization of Citrus Virologists, Brazil, (IOCV, Riverside, 2000), p. 1
N.G. Iglesias, S.P. Gago-Zachert, G. Costa Robledo, N.M.I. Plata, O. Vera, O. Garu, L.C. Semorile, Virus Genes 36, 199–207 (2008)
D. Fargette, G. Konate, C. Fauquet, E. Muller, M. Peterschmitt, J.M. Thresh, Annu. Rev. Phytopathol. 44, 235–260 (2006)
S.J. Harper, T.E. Dawson, M.N. Pearson, Arch. Virol. 155, 471–480 (2010)
Acknowledgments
This work was supported by Department of Biotechnology, Govt. of India. The authors are grateful to K. L. Manjunath, USDA-ARS, NCGRCD, Riverside, CA 90507 for synthesis of primers for amplification of CTV sequences. We also thank R. K. Jain, HOD, Plant Pathology; Prof. Anupam Varma, National Professor, Y. S. Ahlawat and V. G. Malathi, In-charge, Plant Virology Unit; Director, Indian Agricultural Research Institute, New Delhi for providing facilities and their valuable suggestion. We are thankful to R. M. Gade, PDKV, Akola; K. B. Pun, IARI Regional Station, Kalimpong; V. M. Chavan, IARI Regional Station, Pune; for valuable support with surveys.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Biswas, K.K., Tarafdar, A., Diwedi, S. et al. Distribution, genetic diversity and recombination analysis of Citrus tristeza virus of India. Virus Genes 45, 139–148 (2012). https://doi.org/10.1007/s11262-012-0748-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0748-3