Abstract
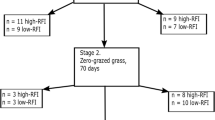
Several measures have been proposed to investigate and improve feed efficiency in cattle. One of the most commonly used measure of feed efficiency is residual feed intake (RFI), which is estimated as the difference between actual feed intake and expected feed intake based on the animal’s average live weight. This measure permits to identify and select the most efficient animals without selecting for higher mature weight. Mitochondrial function has been indicated as a major factor that influences RFI. The analysis of genes involved in mitochondrial function is therefore an alternative to identify molecular markers associated with higher feed efficiency. This study analyzed the expression of PGC1α, TFAM, UCP2 and UCP3 genes by quantitative real-time PCR in liver and muscle tissues of two groups of Nellore cattle divergently ranked on RFI values in order to evaluate the relationship of these genes with RFI. In liver tissue, higher expression of TFAM and UCP2 genes was observed in the negative RFI group. Expression of PGC1α gene did not differ significantly between the two groups, whereas UCP3 gene was not expressed in liver tissue. In muscle tissue, higher expression of TFAM gene was observed in the positive RFI group. Expression of PGC1α, UCP2 and UCP3 genes did not differ significantly between the two groups. These results suggest the use of TFAM and UCP2 as possible candidate gene markers in breeding programs designed to increase the feed efficiency of Nellore cattle.


Similar content being viewed by others
References
Oliveira JS, Zanini AM, Santos EM (2007) Fisiologia, manejo e alimentação de bezerros de corte. Arq. Ciênc. Vet. Zool 1:39–48
Basarab JA (2003) Latest indicator of feed efficiency could spur new generation of efficient cattle. Animal Science newsletter, Flórida. IOP Publishing PhysicsWeb. http://ufdcimages.uflib.ufl.edu/UF/00/06/73/34/00047/00001.pdf. Accessed 11 March 2013
Euclides Filho K, Figueiredo GR, Euclides VPB, Silva LOC, Cusinato VQ (2002) Eficiência bionutricional de animais da raça Nelore seus mestiços com caracu, angus e simental. Rev Bras Zootec 31:331–334
Lanna DPD, Calegari L, Almeida R, Berndt A (2003) Conversão alimentar—eficiência econômica da vacas de corte de raças puras e cruzadas. In: simpósio de pecuária de corte lavras,UFLA, Lavras,87–110
Reunol F (2010) Criações mais eficientes, agência FAPESP, especial. IOP Publishing PhysicsWeb. http://www.agencia.fapesp.br/materia/11704/especiais/criacoes-mais-eficientes. Accessed 5 July 2010
Koch RM, Swinger LA, Chambers D, Gregory KE (1963) Efficiency of feed use in beef cattle. J Anim Sci 22:486–494
Del Claro AC, Mercadante MZ, Silva JA II (2012) Meta-analise de parâmetros genéticos relacionados ao consumo alimentar residual e a suas características componentes em bovinos. Pesqui Agropecu Bras 47:302–310
Zulkifli NA, Naik M, Pitchford WS, Bottema CDK (2007) Cattle residual feed intake candidate genes. J Anim Breed Genet 18:668–671
Bottje W, Tang ZX, Iqbal M, Cawthon D, Okimoto R, Wing T, Cooper M (2002) Association of mitochondrial function with feed efficiency within a single genetic line of male broilers. Poult Sci 81:546–555
Bottje W, Pumford NR, Dirain CO, Iqbal M, Lassiter K (2006) Feed efficiency and mitochondrial function. Poult Sci 85:8–14
Kolath WH, Kerley MS, Golden JW, Keisler DH (2006) The relationship between mitochondrial function and residual feed intake in Angus steers. J Anim Sci 84:861–865
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Gen Biol 3(7):RESEARCH 0034
Steibel JP, Poletto R, Coussens PM, Rosa JMG (2009) A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94:146–152
Ledesma A, Lacoba MG, Rial E (2002) The mitochondrial uncoupling proteins. Gen Biol 3(12):REVIEWS 3015
Echtay KS (2007) Mitochondrial uncoupling proteins—What is their physiological role? Free Radic Biol Med 43:1351–1371
Nordfors L, Hoffstedt J, Nyberg B, Thome A, Arner P, Schalling M (1998) Reduced gene expression of UCP2 but not UCP3 in skeletal muscle of human obese subjects. Diabetologia 41:935–939
Ricquier D, Casteilla L, Bouillaud F (1991) Molecular studies of the uncoupling protein. FASEB J 5:2237–2242
Tsuboyama-Kasaoka N, Ezaki O (2001) Mitochondrial uncopling proteins 3 (UCP3) in skeletal muscle. Front Biosc 6:570–574
Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB (1997) UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun 235:79–82
Kolath WH, Kerley MS, Golden JW, Shahid SA, Johnson GS (2006) The relationships among mitochondrial uncoupling protein 2 and 3 expression, mitochondrial deoxyribonucleic acid single nucleotide polymorphisms, and residual feed intake in Angus steers. J Anim Sci 84:1761–1766
Kelly AK, Waters SM, McGee M, Fonseca RG, Carberry C, Kenny DA (2011) mRNA expression of genes regulating oxidative phosphorylation in the muscle of beef cattle divergently ranked on residual feed intake. Physiol Genomics 43:12–23
Ojano-Dirain C, Toyomizu M, Wing T, Cooper M, Bottje WG (2007) Gene expression in breast muscle and duodenum from low and high feed efficient broilers. Poult Sci 86:372–381
Millet L, Vidal H, Andrielli F, Larrouy D, Rion JP, Ricquier D (1997) Increased uncoupling protein-2 and-3 mRNA expression during fasting in obese and lean humans. J Clin Invest 100:2665–2670
Harrold JA, Widdowson PS, Clapham JC, Williams G (2000) Individual severity of obesity in unselected Wistar rats: relationship with hyperphagia. Am J Physiol 279(2):340–347
Basarab JA, Price MA, Aalhus JL, Okine EK, Snelling WM, Lyle KL (2003) Residual feed intake and body composition in young growing cattle. Can J Anim Sci 83:189–204
Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D (2002) Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA binding protein. EMBO Rep 3:451–456
Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D (2003) Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31:1640–1645
Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, Kim SH, Hamasaki N, Kang D (2007) The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J Biochem 141:201–211
Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson N (2004) Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13:935–944
Gaspari M, Larsson NG, Gustafsson CM (2004) The transcription machinery in mammalian mitochondria. Biochim Biophys Acta 1659:148–152
Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet 31:289–294
Fisher RP, Clayton DA (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol Biol Cell 8:3496–3509
Goffart S, Wiesner RJ (2003) Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp Physiol 88:33–40
Hock MB, Kralli A (2009) Transcriptional control of mitochondrial biogenesis and function. Ann Rev Biochem 7:177–203
Bottje W, Carstens GE (2009) Association of mitochondrial function and feed efficiency in poultry and livestock species. J Anim Sci 87:48–63
Knutti D, Kralli (2001) A PGC-1, a versatile coactivator. Trends Endocrinol Metab 12:360–365
Acknowledgments
This study was supported by the state funding agency São Paulo Research Foundation FAPESP (grants 2009/16118-5 and 2010/13502-6). We thank the Instituto de Zootecnia for providing the tissue samples and database used in this study. We are also grateful to Departamento de Tecnologia, Laboratório de Bioquímica e Biologia Molecular and Programa de Pós-graduação em Genética e Melhoramento Animal, Faculdade de Ciências Agrárias e Veterinárias for their cooperation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fonseca, L.F.S., Gimenez, D.F.J., Mercadante, M.E.Z. et al. Expression of genes related to mitochondrial function in Nellore cattle divergently ranked on residual feed intake. Mol Biol Rep 42, 559–565 (2015). https://doi.org/10.1007/s11033-014-3801-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3801-6