Abstract
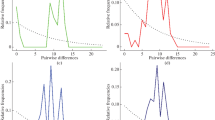
Mitochondrial DNA (mtDNA) D-loop sequences of 666 individuals (including 109 new individuals, 557 individuals retrieved from GenBank) from 33 Chinese domestic goat breeds throughout China were used to investigate their mtDNA variability and molecular phylogeography. The results showed that all goat breeds in this study proved to be extremely diverse, and the average haplotype diversity and nucleotide diversity were 0.990 ± 0.001 and 0.032 ± 0.001, respectively. The 666 sequences gave 326 different haplotypes. Phylogenetic analyses revealed that there were 4 mtDNA haplogroups identified in Chinese domestic goats, in which haplogroup A was predominant and widely distributed. Our finding was consistent with archaeological data and other genetic diversity studies. Amova analysis showed there was significant geographical structuring. Almost 84.31 % of genetic variation was included in the within-breed variance component and only 4.69 % was observed among the geographic distributions. This genetic diversity results further supported the previous view of multiple maternal origins of Chinese domestic goats, and the results on the phylogenetic relationship contributed to a better understanding of the history of goat domestication and modern production of domestic goats.



Similar content being viewed by others
References
FAO (2012) [EB/OL]. http://faostat.fao.org/site/573/default.aspx#ancor. Updated 7 Feb 2014
Chen SY, Su YH, Wu SF, Sha T, Zhang YP (2005) Mitochondrial diversity and phylogeographic structure of Chinese domestic goats. Mol Phylogenet Evol 37(3):804–814. doi:10.1016/j.ympev.2005.06.014
Zhao Y, Zhang JH, Zhao EH, Zhang X, Zhang N XL (2011) Mitochondrial DNA diversity and origins of domestic goats in Southwest China (excluding Tibet). Small Rumin Res 95(1):40–47. doi:10.1016/j.smallrumres.2010.09.004
Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P (2001) Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci USA 98(10):5927–5932
Joshi MB, Rout PK, Mandal AK, Tyler-Smith C, Singh L, Thangaraj K (2004) Phylogeography and origin of Indian domestic goats. Mol Biol Evol 21(3):454–462. doi:10.1093/molbev/msh038
Sultana S, Mannen H, Tsuji S (2003) Mitochondrial DNA diversity of Pakistani goats. Anim Genet 34(6):417–421
Lin BZ, Odahara S, Ishida M, Kato T, Sasazaki S, Nozawa K, Mannen H (2013) Molecular phylogeography and genetic diversity of East Asian goats. Anim Genet 44(1):79–85. doi:10.1111/j.1365-2052.2012.02358.x
Naderi S, Rezaei HR, Taberlet P, Zundel S, Rafat SA, Naghash HR, El-Barody MAA, Ertugrul O, Pompanon F, Econogene C (2007) Large-scale mitochondrial DNA analysis of the domestic goat reveals six haplogroups with high diversity. PLoS One 2(10):e1012. doi:10.1371/journal.pone.0001012
Liu R-Y, Yang G-S, Lei C-Z (2006) The genetic diversity of mtDNA D-loop and the origin of Chinese goats. Acta Genet Sin 33(5):420–428
Zhang H, Li L, Li X (2006) Variability of the complete mitochondrial control region and phylogeny of Sichuan black goats. Anim Biotechnol Bull 10(1):488–493
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 271–275
Parma P, Pietro P, Feligini M, Maria F, Greeppi G, Gianfranco G, Enne G, Giuseppe E (2003) The complete nucleotide sequence of goat (Capra hircus) mitochondrial genome. Goat mitochondrial genome. DNA Seq 14(3):199–203
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599. doi:10.1093/molbev/msm092
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. doi:10.1093/bioinformatics/btp187
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925
Pereira F, Pereira L, Van Asch B, Bradley DG, Amorim A (2005) The mtDNA catalogue of all Portuguese autochthonous goat (Capra hircus) breeds: high diversity of female lineages at the western fringe of European distribution. Mol Ecol 14(8):2313–2318. doi:10.1111/j.1365-294X.2005.02594.x
Li MH, Zhao SH, Bian C, Wang HS, Wei H, Liu B, Yu M, Fan B, Chen SL, Zhu MJ, Li SJ, Xiong TA, Li K (2002) Genetic relationships among twelve Chinese indigenous goat populations based on microsatellite analysis. Genet Sel Evol 34(6):729–744. doi:10.1051/gse:2002032
Naderi S, Rezaei HR, Pompanon F, Blum MGB, Negrini R, Naghash HR, Balkiz O, Mashkour M, Gaggiotti OE, Ajmone-Marsan P, Kence A, Vigne JD, Taberlet P (2008) The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc Natl Acad Sci USA 105(46):17659–17664. doi:10.1073/pnas.0804782105
Agha SH, Pilla F, Galal S, Shaat I, D’Andrea M, Reale S, Abdelsalam AZA, Li MH (2008) Genetic diversity in Egyptian and Italian goat breeds measured with microsatellite polymorphism. J Anim Breed Genet 125(3):194–200
Li X-L, Valentini A (2004) Genetic diversity of Chinese indigenous goat breeds based on microsatellite markers. J Anim Breed Genet 121(5):350–355
Jia Y-H, Shi X-W, Jian C-S, Zhu W-S, Zhang Y-P, He Z-Q, Liao Z-L, Yu Y-H, Li T-Q (1999) Mitochondrial DNA polymorphism of Guizhou goat breeds. Zool Res 20(2):88–92
Li X, Zhang Y, Chen S, Zeng F, Qiu X, Liu X (1997) Study on the mtDNA RFLP of goat breeds. Zool Res 18(4):421–428
Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, Chen W, Chen J, Zeng P, Hou Y, Bian C, Pan S, Li Y, Liu X, Wang W, Servin B, Sayre B, Zhu B, Sweeney D, Moore R, Nie W, Shen Y, Zhao R, Zhang G, Li J, Faraut T, Womack J, Zhang Y, Kijas J, Cockett N, Xu X, Zhao S, Wang J, Wang W (2013) Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol 31(2):135–141. doi:10.1038/nbt.2478
Liu YP, Cao SX, Chen SY, Yao YG, Liu TZ (2009) Genetic diversity of Chinese domestic goat based on the mitochondrial DNA sequence variation. J Anim Breed Genet 126(1):80–89. doi:10.1111/j.1439-0388.2008.00737.x
Wu Y, Guan W, Zhao Q, He X, Pu Y, Ma Y (2008) Mitochondrial DNA genetic diversity of 10 goat breeds. J Agric Biotechnol 16(2):253–257
Wang J, Chen YL, Wang XL, Yang ZX (2008) The genetic diversity of seven indigenous Chinese goat breeds. Small Rumin Res 74(1–3):231–237. doi:10.1016/j.smallrumres.2007.03.007
Liu RY, Lei CZ, Liu SH, Yang CS (2007) Genetic diversity and origin of Chinese domestic goats revealed by complete mtDNA D-loop sequence variation. Asian–Australasian J Anim Sci 20(2):178–183
Royo LJ, Traore A, Tamboura HH, Alvarez I, Kabore A, Fernandez I, Ouedraogo-Sanou G, Toguyeni A, Sawadogo L, Goyache F (2009) Analysis of mitochondrial DNA diversity in Burkina Faso populations confirms the maternal genetic homogeneity of the West African goat. Anim Genet 40(3):344–347. doi:10.1111/j.1365-2052.2008.01828.x
Kul BC, Ertugrul O (2011) mtDNA diversity and phylogeography of some Turkish native goat breeds. Ankara Universitesi Veteriner Fakultesi Dergisi 58(2):129–134. doi:10.1501/Vetfak_0000002462
Cai X, TserangDonko M, Gou X-N, Dong S, Liu C, Wang X (2012) Goats from Sichuan, Chongqing and Guizhou were originated from A and B maternal lineages. Sichuan Daxue Xuebao (Ziran Kexueban) 49(4):887–894. doi:10.3969/j.issn.0490-6756.2012.04.031
Takada T, Kikkawa Y, Yonekawa H, Kawakami S, Amano T (1997) Bezoar (Capra aegagrus) is a matriarchal candidate for ancestor of domestic goat (Capra hircus): evidence from the mitochondrial DNA diversity. Biochem Genet 35(9–10):315–326
Manceau V, Crampe JP, Boursot P, Taberlet P (1999) Identification of evolutionary significant units in the Spanish wild goat, Capra pyrenaica (Mammalia, Artiodactyla). Anim Conserv 2(1):33–39. doi:10.1111/j.1469-1795.1999.tb00046.x
Pidancier N, Jordan S, Luikart G, Taberlet P (2006) Evolutionary history of the genus Capra (Mammalia, Artiodactyla): discordance between mitochondrial DNA and Y-chromosome phylogenies. Mol Phylogenet Evol 40(3):739–749. doi:10.1016/j.ympev.2006.04.002
Fan B, Chen SL, Kijas JH, Liu B, Yu M, Zhao SH, Zhu MJ, Xiong TA, Li K (2007) Phylogenetic relationships among Chinese indigenous goat breeds inferred from mitochondrial control region sequence. Small Rumin Res 73:262–266. doi:10.1016/j.smallrumres.2006.12.007
Chen SL, Fan B, Liu B, Yu M, Zhao SH, Zhu MJ, Xiong T, Li K (2006) Genetic variations of 13 indigenous Chinese goat breeds based on cytochrome b gene sequences. Biochem Genet 44(3–4):89–99. doi:10.1007/s10528-006-9013-6
Azor PJ, Monteagudo LV, Luque M, Tejedor MT, Rodero E, Sierra I, Herrera M, Rodero A, Arruga MV (2005) Phylogenetic relationships among Spanish goats breeds. Anim Genet 36(5):423–425. doi:10.1111/j.1365-2052.2005.01327.x
Amills M, Ramirez O, Tomas A, Badaoui B, Marmi J, Acosta J, Sanchez A, Capote J (2009) Mitochondrial DNA diversity and origins of South and Central American goats. Anim Genet 40(3):315–322. doi:10.1111/j.1365-2052.2008.01837.x
Hou L, Wang J, Li P, Li J, Xing F (2008) MtDNA D-loop polymorphism and phylogeny of sympatric goat populations in lower valley of Yellow river. Chin J Vet Sci 28(12):1474–1479
Acknowledgments
The work was supported by: the National Natural Science Foundation of China (No. 31172195), the Fundamental Research Funds for the Central Universities (No. XDJK2010C092, the 2013 Innovation Team Building Program in Chongqing universities and Natural Science Foundation Project of CQ CSTC (No. CSTC, 2011BB1015).
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, Y., Zhao, R., Zhao, Z. et al. Genetic diversity and molecular phylogeography of Chinese domestic goats by large-scale mitochondrial DNA analysis. Mol Biol Rep 41, 3695–3704 (2014). https://doi.org/10.1007/s11033-014-3234-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3234-2