Abstract
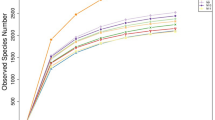
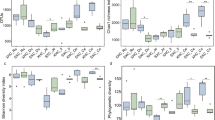
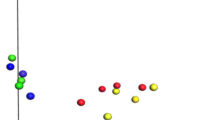
The dominant rumen bacteria in Gayals, Yaks and Yunnan Yellow Cattle were investigated using PCR-DGGE approach. The analysis of DGGE profiles, identification of dominant bands and phylogenetic analysis 16S rDNA sequences in DGGE profiles were combined to reveal the dominant bacterial communities and compared the differences between those cattle species. DGGE profiles revealed that Gayals had the most abundant dominant bacteria and the lowest similarity of intraspecies between individuals than other two cattle species. A total of 45 sequences were examined and sequence similarity analysis revealed that Gayals had the most sequences appeared to uncultured bacteria, accounting for 85.0% of the total sequences, Yaks and Yunnan Yellow Cattle had 44.4 and 68.8% uncultured bacterial sequences, respectively. According to phylogenetic analysis, the rumen dominant bacteria of Gayals were mainly phylogenetically placed within phyla firmicutes and bacteroidetes, and the known bacteria were mainly belonged to the genera Lachnospiraceae bacterium, Ruminococcus flavefaciens and Clostridium celerecrescens. Moreover, the dominant bacteria of Yaks were also mainly belonged to phyla firmicutes and bacteroidetes, and the known dominant bacteria were including Ruminococcus flavefaciens, Butyrivibrio fibrisolvens, Pseudobutyrivibrio ruminis, Schwartzia succinivorans and Clostridiales bacterium, most of them are common rumen bacteria. In addition, the dominant bacteria in Yunnan Yellow Cattle were belonged to phyla firmicutes, bacteroidetes and Actinobacteria, and the known dominant bacteria containing Prevotella sp., Staphylococci lentus, Staphylococcus xylosus and Corynebacterium casei. Present study first detected Staphylococcus lentus and Staphylococcus xylosus in the rumen of cattle.





Similar content being viewed by others
References
Ogimoto K, Imai S (1981) Atlas of rumen microbiology. Tokyo, Japan, p 71
Koike S, Handa Y, Goto H, Sakai K, Miyagawa E, Matsui H, Ito S, Kobayashi Y (2010) Molecular monitoring and isolation of previously uncultured bacterial strains from the sheep rumen. Appl Environ Microbiol 76:1887–1894
Tajima K, Aminov RI, Nagamine T, Ogata K, Nakamura M, Matusi H, Benno Y (1999) Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol Ecol 29:159–169
Wanapat M, Cherdthong A (2009) Use of real-time PCR technique in studying rumen cellulolytic bacteria population as affected by level of roughage in swamp buffalo. Curr Microbiol 58:294–299
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Kocherginskaya SA, Aminov RI, White BA (2001) Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistically ecology approaches. Anaerobe 7:119–134
Konstantinov SR, Zhu WY, Williams BA, Tamminga S, de Vos WM, Akkermans ADL (2003) Effect of fermentable carbohydrates on piglet faecal bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA. FEMS Microbiol Ecol 43:225–235
Sun YZ, Mao SY, Yao W, Zhu WY (2008) DGGE and 16S rDNA analysis reveals a highly diverse and rapidly colonising bacterial community on different substrates in the rumen of goats. Animal 3:391–398
Li SP, Chang H, Ma GL, Cheng HY (2008) Molecular phylogeny of the gayal in Yunnan China inferred from the analysis of cytochrome b gene entire sequences. Asian-australas J Anim Sci 21:789–793
Mondal M, Dhali A, Rajkhowa C, Prakash BS (2004) Secretion patterns of growth hormone in growing captive mithuns (Bos frontalis). Zool Sci 21:1125–1129
Yang SL, Ma SC, Chen J, Mao HM, He YD, Xi DM, Yang LY, He TB, Deng WD (2010) Bacterial diversity in the rumen of Gayals (Bos frontalis), Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences. Mol Biol Rep 37:2063–2073
Mao HM, Deng WD, Wen JK (2005) The biology characteristics of gayals (Bos frontalis) and potential exploitation and utilization. J Yunnan Agric Univ 20:258–261 (in Chinese, with English abstract)
Xi DM, Wanapat M, Deng WD, He TB, Yang ZF, Mao HM (2007) Comparison of Gayal (Bos frontalis) and Yunnan Yellow Cattle (Bos taurus): in vitro dry matter digestibility and gas production for a range of forages. Asian-australas J Anim Sci 20:1208–1214
Deng WD, Wanapat M, Ma SC, Chen J, Xi DM, He TB, Yang ZF, Mao HM (2007) Phylogenetic analysis of 16S rDNA sequences manifest rumen bacterial diversity in Gayals (Bos frontalis) fed fresh bamboo leaves and twigs (Sinarumdinaria). Asian-australas J Anim Sci 20:1057–1066
Deng WD, Wang LP, Ma SC, Jin B, He TB, Yang ZF, Mao HM, Wanapat M (2007) Comparison of Gayal (Bos frontalis) and Yunnan Yellow Cattle (Bos Taurus): rumen function, digestibilities and nitrogen balance during feeding of pelleted Lucerne (Medicago sativum). Asian-australas J Amin Sci 20:900–907
An D, Dong X, Dong Z (2005) Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 4:207–215
Cai L (1995) The Yak. Bangkok, Thailand, pp 37–56
Yang LY, Chen J, Cheng XL, Xi DM, Yang SL, Deng WD, Mao HM (2009) Phylogenetic analysis of 16S rDNA sequences reveals rumen bacterial diversity in Yaks (Bos grunniens). Mol Biol Rep 37:553–562
Stahl DA, Flesher B, Mansfield HR, Montgomery L (1988) Use of phylogenetically based hybridization probes for studies in ruminal microbial ecology. Appl Environ Microbiol 154:1079–1084
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Farris RJ, Garrity GM, Olsen G, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Regensbogenova M, Pristas P, Javorsky P, Moon-Van Der Staay SY, Van Der Staay GWM, Hackstein JHP, Newbold CJ, McEwan NR (2004) Assessment of ciliates in the sheep rumen by DGGE. Lett Appl Microbiol 39:144–147
Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL (2004) Development of an assay to quantify rumen ciliate protozoal biomass in cows using Real-Time PCR. J Nutr 134:3378–3384
Fromin N, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez K, Forestier N, Teyssier-Cuvelle S, Gillet F, Aragno M, Rossi P (2002) Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ Microbiol 4:634–643
Muyze G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Sadet S, Martin C, Meunier B, Morgavi DP (2007) PCR-DGGE analysis reveals a distinct diversity in the bacterial population attached to the rumen epithelium. Animal 1:939–944
Edward JE, McEwan NR, Travis AJ, Wallace RJ (2004) 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86:263–281
Kobayashi Y (2006) Inclusion of novel bacteria in rumen microbiology: need for basic and applied science. Anim Sci J 77:375–385
Flint HJ, Mcpherson EC, Bisset J (1989) Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and β(1-3,1-4) glucanase activities. Appl Environ Microbiol 55:1230–1233
Kirby J, Martin JC, Daniel AS, Flint HJ (1997) Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett 149:213–219
Wina E, Muetzel S, Becker K (2006) The dynamics of major fibrolytic microbes and enzyme activity in the rumen in response to short- and long-term feeding of Sapindus rarak saponins. J Appl Microbiol 100:114–122
Bryant MP, Small N (1956) Characteristics of two new genera of anaerobic curved rods isolated from the rumen of cattle. J Bacteriol 72:16–21
Braune A, Gutshow M, Engst W, Blaut M (2001) Degradation of quercetin and luteolin by Eubacterium ramulus. Appl Environ Microbiol 67:5558–5567
Palop MLL, Valles S, Pinaga F, Flors A (1989) Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium celerecrescens sp. nov. Int J Syst Bacteriol 39:68–71
Forster RJ, Teather RM, Gong J, Deng SJ (1996) 16S rDNA analysis of Butyrivibrio fibrisolvens: phylogenetic position and relation to butyrate-producing anaerobic bacteria from the rumen of white-tailed deer. Lett Appl Microbiol 23:218–222
Stewart CS, Flint HJ, Bryant MP (1997) The rumen bacteria. In: Hobson PN, Stewart CS (eds) The rumen microbial ecosystem, 2nd edn. Chapman and Hall, London, pp 10–72
Tajima K, Arai S, Ogata K, Nagamine T, Matsui H, Nakamura M, Aminov RI, Benno Y (2000) Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe 6:273–284
Gylswyk NO, Hippe H, Rainey FA (1997) Schwartzia succinivorans gen. nov., sp. nov., another ruminal bacterium utilizing succinate as the sole energy source. Int J Syst Bacteriol 47:155–159
Griswold KE, White BA, Mackie RI (1999) Diversity of extracellular proteolytic activities among Prevotella species from the rumen. Curr Microbiol 39:187–194
Wen Z, Morrison M, Wen ZZ (1997) Glutamate dehydrogenase activity profiles for type strains of ruminal Prevotella spp. Appl Environ Microbiol 63:3314–3317
Brennan NM, Brown R, Goodfellow M, Ward AC, Beresford TP, Simpson PJ, Fox PF, Cogan TM (2001) Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 51:843–852
Dehority BA, Tirabasso PA (1998) Effect of ruminal cellulolytic bacterial concentrations on in situ digestion of forage cellulose. J Anim Sci 76:2905–2911
Mackie RI, Aminov RI, Hu WP, Klieve AV, Ouwerkerk D, Sundset MA, Kamagata Y (2003) Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol 69:6808–6815
Wanapat M, Ngarmsang A, Korkhuntot S, Nontaso N, Wachirapakorn C, Beakes G, Rowlinson P (2000) A comparative study on the rumen microbial population of cattle and swamp buffalo raised under traditional village conditions in the Northeast of Thailand. Asian-australas J Anim Sci 13:918–921
Latham MJ, Brooker BE, Pettipher GL, Harris PJ (1978) Adhesion of Bacteroides succinogenes in pure culture and in the presence of Ruminococcus flavefaciens to cell walls in leaves of perennial ryegrass (Lolium perenne). Appl Environ Microbiol 35:1166–1173
Koike S, Yoshitani S, Kobayashi Y, Tanaka K (2003) Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol Lett 229:23–30
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31060314), Yunnan Natural Science Foundation (Grant No. 2010C0038M) and the ‘‘863’’ Key Program of China (Grant No. 2008AA101001) are acknowledged with gratitude.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jing Leng and Linjun Xie contributed equally to this study.
Rights and permissions
About this article
Cite this article
Leng, J., Xie, L., Zhu, R. et al. Dominant bacterial communities in the rumen of Gayals (Bos frontalis), Yaks (Bos grunniens) and Yunnan Yellow Cattle (Bos taurs) revealed by denaturing gradient gel electrophoresis. Mol Biol Rep 38, 4863–4872 (2011). https://doi.org/10.1007/s11033-010-0627-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0627-8