Abstract
Objectives
To alter DNA binding specificity of Vibrio fischeri LuxR and to expand the toolbox for constructing synthetic networks.
Results
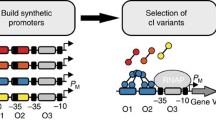
A mutation library (about 10,000 individuals) of the DNA binding domain of LuxR were generated. A genetic selection was performed to obtain LuxR mutants that recognize three lux box DNA variants that are not recognized by wild-type LuxR. Six LuxR mutants were identified. The evolved LuxR mutants were further characterized by measuring the transcriptional activities of different combinations of LuxR mutants and lux box variants. Varied transcriptional activities were found in these LuxR–lux box pairs. The background expressions of the evolved LuxR–lux box systems are more tightly regulated than the wild-type LuxR–lux box system.
Conclusion
The LuxR transcriptional system was evolved to recognize three lux box DNAs which are not recognized by wild-type LuxR.



Similar content being viewed by others
References
Antunes LC, Ferreira RB, Lostroh CP, Greenberg EP (2008) A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397
Balagadde FK et al (2008) A synthetic Escherichia coli predator–prey ecosystem. Mol Syst Biol 4:187
Collins CH, Leadbetter JR, Aronld FH (2006) Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol 24:708–712
Dougherty MJ, Arnold FH (2009) Directed evolution: new parts and optimized function. Curr Opin Biotechnol 20:486–491
Du J, Yuan Y, Si T, Lian J, Zhao H (2012) Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res 40:e142
Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M (2005) The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 151:3589–3602
Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli S, De Francesco R, Cortese R (2003) A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep 4:159–165
Salis H, Tamsir A, Viogt C (2009) Engineering bacterial signals and sensors. Contrib Microbiol 16:194–225
Vannini A, Volpari C, Gargioli C et al (2002) The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J 21:4393–4401
Wei YH, Lai HC, Chen SY, Yeh MS, Chang JS (2004) Biosurfactant production by Serratia marcescens SS-1 and its isogenic strain SMdeltaR defective in SpnR, a quorum-sensing LuxR family protein. Biotechnol Lett 26:799–802
Williams TC, Nielsen LK, Vicker CE (2013) Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol 2:136–149
Wu F, Menn DJ, Wang X (2014) Quorum-sensing cross-talk-driven synthetic circuits: from unimodality to trimodality. Chem Biol 21:1629–1638
Acknowledgments
This work was supported by National High Technology Research and Development Program of China (2012AA02A704) and the Fundamental Research Funds for the Central Universities (Grant No. WK2070000019).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, Y. Engineering Vibrio fischeri transcriptional activator LuxR for diverse transcriptional activities. Biotechnol Lett 38, 1459–1463 (2016). https://doi.org/10.1007/s10529-016-2134-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2134-z