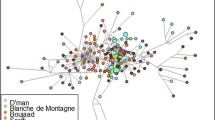
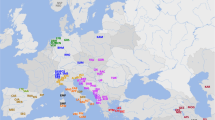
Variations of four sheep populations in China were examined by multiloci electrophoresis, and similar data are quoted to analyze the degree of genetic differentiation of native sheep populations in East and South Asia. Among 15 populations, the average heterozygosity is 0.2746, and the effective number of alleles is 1.559. Mongolian sheep possess the highest genetic diversity, and diversity decreases sequentially in the Chinese, Vietnamese, Bangladeshi, and Nepalese populations. Coefficients of genetic differentiation are 0.0126–0.3083, with an average of 0.148, demonstrating that the major genetic variation (85%) exists within populations. Genetic identity and genetic distance all show relatively low genetic differentiation. No relationship was found between geographic distance and genetic distance. Gene flow is common among the mass of populations, which leads to the inconsistency between geographic distance and genetic distance. The 15 native sheep populations in East and South Asia can be divided into two groups, one group including part of the Chinese and Mongolian populations and another including theYunnan population of China and part of the Nepalese and Bangladeshi populations. Other populations did not separate into groups, merging instead into the two main groups.



Similar content being viewed by others
REFERENCES
Chang, H. (1998). Study on Animal Genetic Resources of China, Shaanxi People's Education, Xi’an, P.R. China, pp. 45–51 (in Chinese).
Geng, R. Q., Chang, H., Yang, Z. P., Sun, W., Wang, L. P., and Lu, S. X. (2002). Study on the genetic investigation and appearance character of Hu sheep. J. Yangzhou Univ. (Agric. Life Sci. edn.) 23(3):37–40 (in Chinese).
Geng, R. Q., Chang, H., Yang, Z. P., Sun, W., Wang, L. P., Lu, S. X., Tsunoda, K., and Ren, Z. J. (2003). Study on origin and phylogeny status of Hu sheep. Asian-Aust. J. Anim. Sci. 16(5):743–747.
Lu, S. X., Chang, H., Du, L., Tsunoda, K., Sun, W., Yang, Z. P., Chang, G. B., and Ji, D. J. (2005a). Phylogenetic relationships of sheep populations from coastal areas in East Asia. Biochem. Genet. 43(5):251–259.
Lu, S. X., Chang, H., Tsunoda, K., Ren, Z. J., Sun, W., Yang, Z. P., Ren, X. L., and Chang, G. B. (2005b). The levers of genetic differentiation of small-tailed Han sheep and Tan sheep populations using structural loci. Sci. Agric. Sin. 38(9):1890–1897 (in Chinese).
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590.
Raymond, M., and Rousset, F. (1995). An exact test for population differentiation. Evolution 49:1280–1283.
Slatkin M. (1987). Gene flow and the geographic structure of natural populations. Science 236:787–792.
Sun, W., Chang, H., Ren, Z. J., Yang, Z. P., Geng, R. Q., Lu, S. X., Du, L., and Tsunoda, K. (2003). Multivariate statistic analysis of morphology and ecology characters on some sheep populations in China. Agric. Sci. in China 11:1271–1276.
Sun, W., Chang, H., Yang, Z. P., Geng, R. Q., Lu, S. X., Chang, G. B., Xu, W., Wang, H. Y., Du, L., Ren, Z. J., and Tsunoda, K. (2002). Studies on the genetic relationships of sheep populations from East and South of Central Asia. Asian-Aust. J. Anim. Sci. 15(10):1398–1402.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595.
Tsunoda, K., Amano, T., and Nozawa, K. (1990). Genetic characteristics of Bangladeshi sheep as based on biochemical variations. Jpn. J. Zootech. Sci. 61(1):54–66.
Tsunoda, K., Doge, K., and Yamamoto, Y. (1992). Morphological traits and blood protein variation of the native Nepalese sheep. Rep. Soc. Res. Native Livestock 14:155–183.
Tsunoda, K., Nozawa, K., and Maeda, Y. (1999). External morphological characters and blood protein and nonprotein polymorphisms of native sheep in Central Mongolia. Rep. Soc. Res. Native Livestock 17:63–82.
Tsunoda, K., Nozawa, K., and Okamoto, S. (1995). Blood protein variation of native sheep populations in Lufeng and Lunan in Yunnan province of China. Rep. Soc. Res. Native Livestock 15:119–129.
Tsunoda, K., Okabayashi, H., and Amano, T. (1998). Morphologic and genetic characteristic of sheep raised by the Cham Tribe in Vietnam. Rep. Soc. Res. Native Livestock 16:63–73.
Wang, M. N., and Li, Z. Q. (1991). Evolution of Animal, Nanjing University Press, Nanjing, P.R. China, pp. 71–78 (in Chinese).
Wright, S. (1931). Evolution in Mendelian population. Genetics 16:91–159.
Yang, Z. P., Chang, H., Sun, W., Geng, R. Q., and Tsunoda, K. (2004). The genetic diversity of some sheep populations in China and the East and South of Central Asia. Acta Agric. Boreali-Occidentalis Sin. 13(2):1–6 (in Chinese).
ACKNOWLEDGMENTS
This research was supported by the International Cooperation Project of the National Natural Science Foundation of P. R. China (Grant nos. 30213001, 30210103007, and 3041013150) and the Project of the Basic Natural Science Foundation for Colleges and Universities in Jiangsu Province (06KJD230203).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Geng, R., Chang, H., Wang, L. et al. Genetic Differentiation of Native Sheep Populations in East and South Asia. Biochem Genet 45, 263–279 (2007). https://doi.org/10.1007/s10528-006-9073-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-006-9073-7