Abstract
Osmotic stresses such as drought, salinity, and cold are major environmental factors that limit agricultural productivity. Transcription factors play essential roles in abiotic stress signaling in plants. Three TaMYB2 members were identified and designated TaMYB2A, TaMYB2B, and TaMYB2D based on their genomic origins. The cis-regulatory elements in the promoter regions were compared, and their diverse expression patterns under different abiotic stress conditions were identified. TaMYB2A was further characterized because of its earlier response to stresses. Subcellular localization revealed that TaMYB2A localized in the nucleus. To examine the role of TaMYB2A under various environmental stresses, transgenic Arabidopsis plants carrying TaMYB2A controlled by the CaMV 35S promoter were generated and subjected to severe abiotic stress. TaMYB2A transgenics had enhanced tolerance to drought, salt, and freezing stresses, which were confirmed by the enhanced expressions of abiotic stress-responsive genes and several physiological indices, including decreased rate of water loss, enhanced cell membrane stability, improved photosynthetic potential, and reduced osmotic potential. TaMYB2A is a multifunctional regulatory factor. Its overexpression confers enhanced tolerance to multiple abiotic stresses while having no obvious negative effects on phenotype under well-watered and stressed conditions; thus, TaMYB2A has the potential for utilization in transgenic breeding to improve abiotic stress tolerances in crops.











Similar content being viewed by others
Abbreviations
- AAR:
-
Amino acid residue
- ABA:
-
Abscisic acid
- ABRE:
-
ABA response element
- CMS:
-
Cell membrane stability
- GFP:
-
Green fluorescent protein
- MBS:
-
MYB-binding site
- OA:
-
Osmotic adjustment
- OP:
-
Osmotic potential
- PEG:
-
Polyethylene glycol
- qRT-PCR:
-
Quantitative real-time PCR
- TF:
-
Transcription factor
- VC:
-
Vector control
- WT:
-
Wild type
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78. doi:10.1046/j.1365-313X.2003.01953.x
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645. doi:10.1074/jbc.M605895200
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58. doi:10.1080/07352680590910410
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380. doi:10.1111/j.1365-313X.2004.02138.x
Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7:1099–1111
Bray E (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wendel JF (2003) Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol Biol 51:313–325. doi:10.1023/A:1022051100610
Chaerle L, Leinonen I, Jones HG, Van Der Straeten D (2007) Monitoring and screening plant populations with combined thermal and chlorophyll fluorescence imaging. J Exp Bot 58:773–784. doi:10.1093/jxb/erl257
Chen R, Ni Z, Nie X, Qin Y, Dong G, Sun Q (2005) Isolation and characterization of genes encoding Myb transcription factor in wheat (Triticum aestivum L.). Plant Sci 169:1146–1154. doi:10.1016/j.plantsci.2005.07.018
Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM et al (2006) The MYB transcription factor superfamoly of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124. doi:10.1007/s11103-005-2910-y
Clarke J, Romagosa M, Jana I, Srivastava JP, McCaig TN (1989) Relationship of excised-leaf water loss rate and yield of durum wheat in diverse environments. Can J Plant Sci 69:1075–1081
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200. doi:10.1016/j.cub.2005.05.048
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Dhanda SS, Sethi GS (1998) Inheritance of excised-leaf water loss and relative water content in bread wheat (Triticum aestivum). Euphytica 104:39–47
Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36:17–29. doi:10.1016/S1673-8527(09)60003-5
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763. doi:10.1046/j.1365-313X.2003.01661.x
Farooq S, Azam F (2006) The use of cell membrane stability (CMS) technique to screen for salt tolerant wheat varieties. J Plant Physiol 163:629–637. doi:10.1016/j.jplph.2005.06.006
Finkelstein RR, Somerville CR (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94:1172–1179
Franca MGC, Thi ATP, Pimentel C, Rossiello ROP, Zuily FY, Laffray D (2000) Differences in growth and water relations among Phaseolus vulgaris cultivars in response to induced drought stress. Environm Exp Bot 43:227–237. doi:10.1016/S0098-8472(99)00060-X
Gray J (2005) Guard cells: transcription factors regulate stomatal movements. Curr Biol 15:R593–R595. doi:10.1016/j.cub.2005.07.039
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Hecht A, Stemmler MP (2003) Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J Biol Chem 278:3776–3785. doi:10.1074/jbc.M210081200
Hong SW, Jon JH, Kwak JM, Nam HG (1997) Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopis thaliana. Plant Physiol 113:1203–1212
Hu CA, Delauney AJ, Verma DP (1992) A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA 89:9354–9358
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992. doi:10.1073/pnas.0604882103
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–403
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635. doi:10.1104/pp.107.110981
Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42:313–349
Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20:951–962
Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW (2007) A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiologia Plantarum 129:375–385. doi:10.1111/j.1399-3054.2006.00828.x
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. doi:10.1093/nar/30.1.325
Levitt J (1980) Responses of plants to environmental stresses. Volume I., Water, radiation, salt and other stresses. Academic Press, New York, pp 3–211
Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15:1201–1206. doi:10.1016/j.cub.2005.06.041
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X et al (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256. doi:10.1104/pp.108.133454
Mao X, Zhang H, Tian S, Chang X, Jing R (2010) TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot 61:683–696. doi:10.1093/jxb/erp331
Mattana M, Biazzi E, Consonni R, Locatelli F, Vannini C, Provera S, Coraggio I (2005) Overexpression of Osmyb4 enhances compatible solute accumulation and increases stress tolerance of Arabidopsis thaliana. Physiol Plant 125:212–223. doi:10.1111/j.1399-3054.2005.00551.x
Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119:463–470
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314. doi:10.1073_pnas.0401572101
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Newman LJ, Perazza DE, Juda L, Campbell MM (2004) Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J 37:239–250. doi:10.1046/j.1365-313X.2003.01953.x
Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E (1999) Maize R2R3 Myb genes: sequence analysis reveals amplification in the higher plants. Genetics 153:427–444
Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2:282–291. doi:10.1007/s10142-002-0070-6
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14:194–199. doi:10.1016/S0958-1669(03)00030-2
Shinozaki K, Yamaguchi S (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327–334
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417. doi:10.1016/S1369-5266(03)00092-X
Soderman E, Mattsson J, Engstrom P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and abscisic acid. Plant J 10:375–381
Strasser A, Srivastava A, Tsimilli-Michae ML (2002) The fluorescence transient as a tool to characterize and screen photosynthetic samples. The fluorescence transient as a tool to characterize and screen photosynthetic samples. In: Mohanty P, Yunus U, Pathre M (eds) Probing photosynthesis: mechanism, regulation and adaptation. Taylor and Francis, London, pp 443–480
Takahashi S, Katagiri T, Hirayama T, Yamaguchi-Shinozaki K, Shinozaki K (2001) Hyperosmotic stress induces a rapid and transient increase in inositol 1,4,5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol 42:214–222. doi:10.1093/pcp/pce028
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N (2006) Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet 112:445–454. doi:10.1007/s00122-005-0144-7
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2:135–138
Urao T, Noji M, Yamaguchi-Shinozaki K, Shinozaki K (1996) A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J 10:1145–1148
Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A (1996) Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci 21:59–64
Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37:115–127. doi:10.1046/j.1365-313X.2003.01938.x
Vannini C, Iriti M, Bracale M, Locatelli F, Faoro F, Croce P, Pirona P, Di Maro A, Coraggio I, Genga A (2006) The ectopic expression of the rice Osmyb4 gene in Arabidopsis increases tolerance to abiotic, environmental and biotic stresses. Physiol Mol Plant Pathol 69:26–42. doi:10.1016/j.plantsci.2007.05.007
Vannini C, Campa M, Iriti M, Genga A, Faoro F, Carravieri S, Rotino GL, Rossoni M, Spinardi M, Bracale A (2007) Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci 173:231–239. doi:10.1016/j.gene.2008.06.024
Wang AP, Mao XG, Jing RL, Chang XP, Yang W (2006) Single nucleotide polymorphism of TaMyb2-II gene in common wheat (Triticum aestivum L.) and its relatives. Act Agron Sin 32:1809–1816
Xiong L, Ishitani M, Zhu JK (1999) Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol 119:205–212
Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340
Zhou D, Zhou J, Meng L, Wang Q, Xie H, Guan Y, Ma Z, Zhong Y, Chen F, Liu J (2009) Duplication and adaptive evolution of the COR15 genes within the highly cold-tolerant Draba lineage (Brassicaceae). Gene 441:36–44. doi:10.1016/j.gene.2008.06.024
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. doi:10.1146/annurev.arplant.53.091401.143329
Acknowledgments
The authors thank Prof. Zhensheng Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for providing the pJIT163-GFP expression vector. We thank Robert A. McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for critical reading and comments on the manuscript. This study was supported by the National Science Foundation of China (31040089), Key Project of Chinese National Programs for Fundamental Research and Development (2010CB951501), and the National Key Technologies R&D Program (2009ZX08002-012B).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xinguo Mao and Dongsheng Jia contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
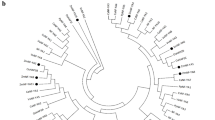
Phylogenetic tree of TaMYB2 genes from related diploid and tetraploid species and hexaploid wheat. The maximum likelihood tree was constructed with the PROML program in the PHYLIP software package, with the bootstrap parameter set at 100. The three TaMYB2 members are underlined (PDF 13 kb)
Fig. S2
Morphological characterization of TaMYB2A plants. A. Comparison of primary root lengths. Arabidopsis seeds were planted on MS plates and cultured vertically, and primary root lengths were compared on the 7th day (F test, P < 0.05). B. Comparison of lateral root numbers on transgenic lines and WT plants (F test, P < 0.05). C. The overexpression of TaMYB2A results in earlier flowering in Arabidopsis (PDF 99 kb)
Fig. S3
Expression levels of TaMYB2A varied widely among different transgenic Arabidopsis lines. L1–L6, individual TaMYB2A transgenic lines (PDF 8 kb)
Table S1
Oligonucleotides for qRT-PCR in wheat (DOC 32 kb)
Rights and permissions
About this article
Cite this article
Mao, X., Jia, D., Li, A. et al. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis . Funct Integr Genomics 11, 445–465 (2011). https://doi.org/10.1007/s10142-011-0218-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0218-3