Abstract
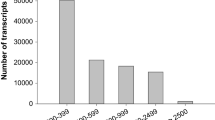
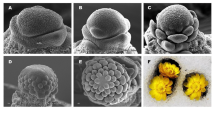
The understanding of flower initiation, development, and maturation in mangosteen is of paramount importance to shorten its long juvenile phase and to synchronize its flowering or fruiting time. In this study, we have identified 97 tentative unique genes with higher expression levels in young flower buds compared to young shoots by using suppressive subtraction hybridization and reverse northern analysis. Sequence analysis showed that 63.9% of these transcripts had non-significant matches to sequences in the non-redundant protein database in GenBank, 19.6% had significant matches to unknown proteins while the remaining 16.5% had putative functions in transcription, stress, signal transduction, cell wall biogenesis, photosynthesis and miscellaneous. The full-length cDNA of GmAGMBP encoding AG-motif binding protein (a zinc finger transcriptional factor), and 3′ termini cDNA sequences of GmHSA32 and GmBZIP, encoding heat-stress-associated 32 (HSA32) and bZIP transcription factor, respectively; were cloned and further analysed. Real-time PCR analysis revealed that these three genes have different transcript profiles in flowers of different developmental stages and young shoots. The highest abundance of transcripts was achieved in flowers with diameters ranging from 0.5 to 0.9 cm for GmAGMBP and GmBZIP and in flowers with diameters less than 0.5 cm for GmHSA32. Southern analysis suggested that GmAGMBP might be single copy gene while GmHSA3A could possibly belong to a small gene family in the mangosteen genome.






Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EM, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1998) Current protocols in molecular biology. Wiley, New York
Cavener DR, Ray SC (1991) Eukaryotic start and stop translation site. Nucleic Acids Res 19:3185–3192. doi:10.1093/nar/19.12.3185
Chan K-L, Ho C-L, Namasivayam P, Napis S (2007) A simple and rapid method for RNA isolation from plant tissues with high phenolic compounds and polysaccharides. Nat Protoc. doi:10.1038/nprot.2007.184
Chang YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabisopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140:1297–1305. doi:10.1104/pp.105.074898
Hsu YF, Wang CS, Raja R (2007) Gene expression pattern at desiccation in the anther of Lilium longiflorum. Planta 226:311–322. doi:10.1007/s00425-007-0483-5
Hu W, Wang YX, Bowers C, Ma H (2003) Isolation, sequence analysis, and expression studies of florally expressed cDNAs in Arabidopsis. Plant Mol Biol 53:545–563. doi:10.1023/B:PLAN.0000019063.18097.62
Huang A, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877. doi:10.1101/gr.9.9.868
Jordan BR, Anthony RG (1993) Floral homeotic genes: isolation, characterization and expression during floral development. In: Jordan BR (ed) The molecular biology of flowering. C.A.B. International, UK, pp 93–116
Joshi CP, Zhou H, Huang X, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35:993–1001. doi:10.1023/A:1005816823636
Kobayashi A, Sakamoto A, Kubo K, Rybka Z, Kanno Y, Takatsuji H (1998) Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J 13:571–576. doi:10.1046/j.1365-313X.1998.00043.x
Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY (2008) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228:225–240. doi:10.1007/s00425-008-0731-3
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \( 2^{\Updelta \Updelta C_{\text{T}} } \)method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Lü S, Gu H, Yuan X, Wang X, Wu AM, Qu L, Liu JY (2007) The GUS reporter-aided analysis of the promoter activities of a rice metallothionein gene reveals different regulatory regions responsible for tissue-specific and inducible expression in transgenic Arabidopsis. Transgenic Res 16:177–191. doi:10.1007/s11248-006-9035-1
Martin J, Storgaard M, Andersen CH, Nielsen KK (2004) Photoperiodic regulator of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol Biol 56:159–169. doi:10.1007/s11103-004-2647-z
Matsumoto TK (2006) Genes uniquely expressed in vegetative and potassium chlorate induced floral buds of Dimocarpus longan. Plant Sci 170:500–510. doi:10.1016/j.plantsci.2005.09.016
Nemoto Y, Kisaka M, Ogihara Y (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J 36:82–93. doi:10.1046/j.1365-313X.2003.01859.x
Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64:1402–1409. doi:10.1271/bbb.64.1402
Ong HT (1976) Climatic changes in water balance and their effects on tropical flowering. Planter 52:174–179
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203. doi:10.1038/35012103
Pelaz S, Tapia-Lopez R, Alvarez-Buylla ER, Yanofsky MF (2001) Conversion of leaves into petals in Arabidopsis. Curr Biol 11:182–184. doi:10.1016/S0960-9822(01)00024-0
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Res 29:e45. doi:10.1093/nar/29.9.e45
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857. doi:10.1016/0092-8674(95)90288-0
Rozen S, Skaletsky HJ (2000) Primer 3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics method and protocols: methods in molecular biology. Human Press, Totowa, pp 365–386
Sarowar S, Oh HW, Cho HS, Baek KH, Seong ES, Joung YH, Choi GJ, Lee S, Choi D (2007) Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 51:792–802. doi:10.1111/j.1365-313X.2007.03174.x
Shikata M, Takemura M, Yokota A, Kohchi T (2003) Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci Biotechnol Biochem 67:2459–2497. doi:10.1271/bbb.67.2495
Simon R, Coupland G (1996) Arabidopsis genes that regulate flowering time in response to day-length. Semin Cell Dev Biol 7:419–425. doi:10.1006/scdb.1996.0052
Song J, Yamamoto K, Shomura A, Itadani H, Zhong HS, Yano M, Sasaki T (1998) Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Res 5:95–101. doi:10.1093/dnares/5.2.95
Strathmann A, Kuhlmann M, Heinekamp T, Droge-Laser W (2001) BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TB2F and BZI-4, and is functionally involved in flower development. Plant J 28:397–408. doi:10.1046/j.1365-313X.2001.01164.x
Su PH, Li HM (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146:1231–1241. doi:10.1104/pp.107.114496
Sugimoto K, Takeda S, Hirochika H (2003) Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J 36:550–564. doi:10.1046/j.1365-313X.2003.01899.x
Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. doi:10.1186/1471-2164-8-125
Takaiwa F, Oono K, Sugiura M (1984) The complete nucleotide sequence of a rice 17S rRNA gene. Nucleic Acids Res 12:5441–5448
Takatsuji H, Nakamura N, Katsumoto Y (1994) A new family of zinc finger proteins in petunia: structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell 6:947–958
Theissen G, Melzer R (2007) Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot (Lond) 100:603–619. doi:10.1093/aob/mcm143
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Yaacob O, Tindall HD (1995) Mangosteen cultivation. FAO Plant Production and Protection Paper No. 129
Yuan J, Chen D, Ren Y, Zhang X, Zhao J (2008) Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol 146:1637–1650. doi:10.1104/pp.107.110304
Zhang XH, Feng BM, Zhang Q, Zhang D, Altman N, Ma H (2005) Genome-wide expression profiling and identification of gene activities during early flower development in Arabidopsis. Plant Mol Biol 58:401–419. doi:10.1007/s11103-005-5434-6
Zhao Y, Wang G, Zhang J, Yang J, Peng S, Gao L, Li C, Hu J, Li D, Gao L (2006) Expressed sequence tags (ESTs) and phylogenetic analysis of floral genes from a paleoherb species, Asarum caudigerum. Ann Bot (Lond) 98:157–163. doi:10.1093/aob/mcl081
Zhou YT, Wang HY, Zhou L, Wang MP, Li HP, Wang ML, Zhao Y (2008) Analyses of the floral organ morphogenesis and the differentially expressed genes of an apetalous flower mutant in Brassica napus. Plant Cell Rep 27:9–20. doi:10.1007/s00299-007-0426-4
Acknowledgments
The authors gratefully acknowledge the nursery at Universiti Putra Malaysia, Serdang, Malaysia, for providing the mangosteen tissues, and the assistance of Miss R.-S. Siow in RNA extraction from mangosteen tissues is also acknowledged. This project was funded by the Intensified Research Grant for Priority Area (IRPA) No: 01-03-03-005 BTK/ER/02 from the Ministry of Science, Technology and Innovation (MOSTI) of Malaysia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Canovas.
Rights and permissions
About this article
Cite this article
Chan, KL., Yeoh, KA., Lim, KA. et al. Isolation and characterization of floral transcripts from mangosteen (Garcinia mangostana L.). Trees 23, 899–910 (2009). https://doi.org/10.1007/s00468-009-0331-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-009-0331-2