Abstract
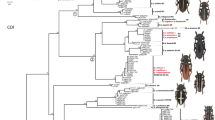
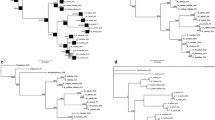
The complete internal transcribed spacer 1 (ITS1), 5.8S ribosomal DNA, and ITS2 region of the ribosomal DNA from 60 specimens belonging to two closely related bucephalid digeneans (Dollfustrema vaneyi and Dollfustrema hefeiensis) from different localities, hosts, and microhabitat sites were cloned to examine the level of sequence variation and the taxonomic levels to show utility in species identification and phylogeny estimation. Our data show that these molecular markers can help to discriminate the two species, which are morphologically very close and difficult to separate by classical methods. We found 21 haplotypes defined by 44 polymorphic positions in 38 individuals of D. vaneyi, and 16 haplotypes defined by 43 polymorphic positions in 22 individuals of D. hefeiensis. There is no shared haplotypes between the two species. Haplotype rather than nucleotide diversity is similar between the two species. Phylogenetic analyses reveal two robustly supported clades, one corresponding to D. vaneyi and the other corresponding to D. hefeiensis. However, the population structures between the two species seem to be incongruent and show no geographic and host-specific structure among them, further indicating that the two species may have had a more complex evolutionary history than expected.



Similar content being viewed by others
References
Anderson GR, Barker SC (1998) Inference of phylogeny and taxonomy within the Didymozoidae (Digenea) from the second internal transcribed spacer (ITS2) of ribosomal DNA. Syst Parasitol 41:87–94
Bell AS, Somerville C, Valtonen ET (2001) A molecular phylogeny of the genus Ichthyocotylurus (Digenea, Strigeidae). Int J Parasitol 31:833–842
Brandley MC, Schmitz A, Reeder TW (2005) Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol 54:373–390
Buckley TR (2002) Model misspecification and probabilistic tests of topology: evidence from empirical data sets. Syst Biol 51:509–523
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Donoghue MJ, Moore BR (2003) Toward an integrative historical biogeography. Integr Comp Biol 43:261–270
Felsensten JP (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Galazzo DE, Dayanandan S, Marcogliese DJ, McLaughlin JD (2002) Molecular systematics of some North American species of Diplostomum (Digenea) based on rDNA sequence data and comparisons with European congeners. Can J Zool 80:2207–2217
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Jermiin LS, Ho SYW, Ababneh F, Robinson J, Larkum AWD (2004) The biasing effect of compositional heterogeneity on phylogenetic estimates may be underestimated. Syst Biol 53:638–643
Johnson KP, Adams RJ, Page RDM, Clayton DH (2003) When do parasites fail to speciate in response to host speciation? Syst Biol 52:37–47
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics 5:150–163
Luo H-Y, Nie P, Zhang Y-A, Wang G-T, Yao W-J (2002) Molecular variation of Bothriocephalus acheilognathi Yamaguti, 1934 (Cestoda: Pseudophyllidea) in different fish host species based on ITS rDNA sequences. Syst Parasitol 52:159–166
Luton K, Walker D, Blair D (1992) Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea). Mol Biochem Parasitol 56:323–328
Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45:524–545
McManus DP, Bowles J (1996) Molecular genetic approaches to parasite identification: their value in diagnostic parasitology and systematics. Int J Parasitol 26:687–704
Michaux JR, Libois R, Filippucci MG (2005) So close and so different: comparative phylogeography of two small mammal species, the yellow-necked fieldmouse (Apodemus xavicollis) and the woodmouse (Apodemus sylvaticus) in the Western Palearctic region. Heredity 94:52–63
Niewiadomska K, Laskowski Z (2002) Systematic relationships among six species of Diplostomum Nordmann, 1832 (Digenea) based on morphological and molecular data. Acta Parasitol 47:1230–1237
Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Page RDM (2003) Tangled trees: phylogeny, cospeciation and coevolution. University of Chicago Press, Chicago
Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55:595–609
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Rocha LA, Robertson DR, Roman J, Bowen BW (2005) Ecological speciation in tropical reef fishes. Proc R Soc Lond B 272:573–579
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DNAsp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Shimodaria H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–598
Stunzenas V, Cryan JR, Molloy DP (2004) Comparison of rDNA sequences from colchicine treated and untreated sporocysts of Phyllodistomum folium and Bucephalus polymorphus (Digenea). Parasitol Int 53:223–228
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer, Sunderland, MA
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tkach VV, Pawlowski J, Sharpilo VP (2000) Molecular and morphological differentiation between species of the Plagiorchis vespertilionis group (Digenea, Plagiorchiidae) occurring in European bats, with a re-description of P. vespertilionis (Muller, 1780). Syst Parasitol 47:9–22
Wang G-T, Wang W-J (1998a) Taxonomy of the family Bucephalidae (Digenea). Acta Hydrobiol Sin 22 (suppl):89–99 (in Chinese)
Wang G-T, Wang W-J (1998b) Taxonomy and keys to the bucephalid species in China, with descriptions of three new species. Acta Hydrobiol Sin 22(suppl):100–110 (in Chinese)
Wang G-T, Wang W-J (2000) The life cycle of Dollfustrema vaneyi. Acta Hydrobiol Sin 24:644–647 (In Chinese)
Zhang J-Y, Qiu, Z-Z, Ding X-J et al (1999) Parasites and parasitic diseases of fishes. Science Press, Beijing, China (in Chinese)
Acknowledgments
The first author thanks Dr. Q. Gao for assistance in collecting specimens. We are grateful to Dr. X. Guo (Chengdu Institute of Biology, Chinese Academy of Sciences) for his kind help on the Bayesian hypothesis testing and helpful comments on multiple drafts of this manuscript. We also gracefully acknowledge the two anonymous reviewers and the editor whose critical review and insightful suggestions have helped to improve the clarity and focus of the manuscript. This study was supported by grants (project no. 30130150) from the National Natural Science Foundation of China and a grant [project no. (2005) 192] from the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, D., Wang, G., Yao, W. et al. Utility of ITS1–5.8S–ITS2 sequences for species discrimination and phylogenetic inference of two closely related bucephalid digeneans (Digenea: Bucephalidae): Dollfustrema vaneyi and Dollfustrema hefeiensis . Parasitol Res 101, 791–800 (2007). https://doi.org/10.1007/s00436-007-0549-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-007-0549-0