Abstract
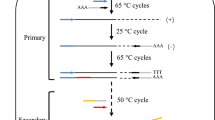
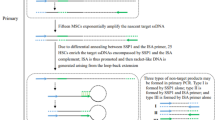
High-efficiency thermal asymmetric interlaced (HE-TAIL) PCR is a modified thermal asymmetric interlaced (TAIL) method for finding unknown genomic DNA sequences adjacent to known sequences in GC-rich plant DNA. Necessary modifications to obtain high-efficiency amplification of flanking sequences are the inclusion of 2 control reactions during tertiary cycling and the design of long gene-specific primers, which can be used during single-step annealing-extension PCR. The modified protocol is suitable to walk from short known sequences, such as sequence-tagged sites (STS), expressed sequence tags (EST), or short exon sequences, and enables researchers to clone full-length open reading frames (ORFs) without library screening. Moreover, the HE-TAIL method can be used to identify DNA sequences flanking T-DNA insertions or to isolate promoter regions. Although individual steps are limited to about 4 kb, multiple steps can be done to walk upstream or downstream of known regions.
Similar content being viewed by others
Abbreviations
- EtBr:
-
ethidium bromide
- HE-TAIL:
-
high-efficiency thermal asymmetric interlaced
References
Cottage A, Yang A, Maunders H, Lacy C, and Ramsay N (2001) Identification of DNA sequences flanking T-DNA insertions by PCR-walking. Plant Mol Biol Rep 19: 321–327.
Dabo SM, Mitchell ED, and Melcher U (1993) A method for isolation of nuclear DNA from cotton leaves. Anal Biochem 210: 34–38.
Dempster EL, Pryor KV, Francis D, Young JE, and Rogers HJ (1999) Rapid DNA extraction from Ferns for PCR based analysis. BioTechniques 27: 66–68.
Liu Y, Mitsukawa N, Oosumi T, and Whittier RF (1995) Efficient isolation and mapping ofArabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463.
Liu Y and Whittier R (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681.
Mazars GR, Moyret C, Jeanteur P, and Theillet CG (1991) Direct sequencing by thermal asymmetric PCR. Nucl Acids Res 19: 4783.
Michiels A, Van den Ende W, Tucker M, Van Riet L, and Van Laere A (2003) Extraction of high-quality genomic DNA from latex-containing plants. Anal Biochem 315: 85–89.
Nakazaki T, Ihara N, Fucuta Y, and Ikehashi H (2000) Abundant polymorphism in the flanking regions of two loci for basic PR-1 proteins as markers forIndica japonica differentiation in rice. Breeding Sci 50: 173–181.
Nakazaki T and Ikehashi H (1998) Genomic sequence and polymorphisms of a rice chitinase gene, Cht4. Breeding Sci 48: 371–376.
Ochman H, Gerber A, and Hartl D (1988) Genetic application of an inverse polymerase chain reaction. Genetics 120: 621–623.
Rosenthal A and Jones D (1990) Genomic walking and sequencing by oligo-cassette mediated polymerase chain reaction. Nucl Acid Res 18: 3095–3096.
Sato Y, Murakami T, Funatsuki H, Matsuba S, Saruyama H, and Tanida M (2001) Heat shock mediated APX gene expression and protection against chilling injury in rice seedlings. J Exp Bot 52: 145–151.
Siebert P, Chenchik A, Kellog D, Lukyanov K, and Lukyanov S (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acid Res 23: 1087–1088.
Terauchi R and Kahl G (2000) Rapid isolation of promoter sequences by TAIL-PCR: the 5′ flanking regions of Pal and Pgi genes from yams (Dioscorea). Mol Gen Genet 263: 554–560.
Van den Ende W, Michiels A, Van Wonterghem D, Clerens S, De Roover J, and Van Laere A (2001) Defoliation induces 1-FEHII (Fructan 1-exohydrolase II) in witloof chicory roots. Cloning and purification two isoforms (1-FEH IIa and 1-FEH IIb). Mass fingerprint of the 1-FEH II enzymes. Plant Physiol 126: 1186–1195.
Woodhead M, Davies HV, Brennan RM, and Taylor MA (1998) The isolation of genomic DNA from Blackcurrant (Ribes nigrum L.). Mol Biotech 9: 243–246.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Michiels, A., Tucker, M., van den Ende, W. et al. Chromosomal walking of flanking regions from short known sequences in GC-rich plant genomic DNA. Plant Mol Biol Rep 21, 295–302 (2003). https://doi.org/10.1007/BF02772805
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF02772805