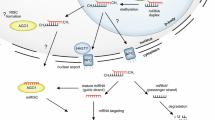
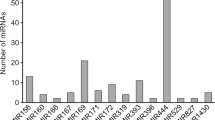
Abstract
MicroRNAs (miRNAs) are a newly discovered class of noncoding endogenous small RNAs involved in plant growth and development as well as response to environmental stresses. miRNAs have been extensively studied in various plant species, however, only few information are available in cassava, which serves as one of the staple food crops, a biofuel crop, animal feed and industrial raw materials. In this study, the 169 potential cassava miRNAs belonging to 34 miRNA families were identified by computational approach. Interestingly, mes-miR319b was represented as the first putative mirtron demonstrated in cassava. A total of 15 miRNA clusters involving 7 miRNA families, and 12 pairs of sense and antisense strand cassava miRNAs belonging to six different miRNA families were discovered. Prediction of potential miRNA target genes revealed their functions involved in various important plant biological processes. The cis-regulatory elements relevant to drought stress and plant hormone response were identified in the promoter regions of those miRNA genes. The results provided a foundation for further investigation of the functional role of known transcription factors in the regulation of cassava miRNAs. The better understandings of the complexity of miRNA-mediated genes network in cassava would unravel cassava complex biology in storage root development and in coping with environmental stresses, thus providing more insights for future exploitation in cassava improvement.





Similar content being viewed by others
References
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
Carthew, R. W., & Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655.
Jones-Rhoades, M. W., Bartel, D. P., & Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annual Review plant Biology, 57, 19–53.
Zhang, B. H., Pan, X. P., Cobb, G. P., & Anderson, T. A. (2006). Plant microRNA: a small regulatory molecule with big impact. Developmental Biology, 289, 3–16.
Dugas, D. V., & Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Current Opinion in Plant Biology, 7, 512–520.
Chen, X. (2005). MicroRNA biogenesis and function in plants. FEBS Letters, 579, 5923–5931.
Cui, X., Xu, S. M., Mu, D. S., & Yang, Z. M. (2009). Genomic analysis of rice microRNA promoters and clusters. Gene, 431, 61–66.
Lee, Y., Kim, M., Han, J., Yeom, K., Lee, S., et al. (2004). MicroRNA genes are transcribed by RNA polymeraseII. EMBO Journal, 23, 4051–4060.
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., & Bartel, D. P. (2002). Micro-RNAs in plants. Genes and Development, 16, 1616–1626.
Papp, I., Mette, M. F., Aufsatz, W., Daxinger, L., Schauer, S. E., et al. (2003). Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiology, 132, 1382–1390.
Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H., & Poethig, R. S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proceedings National Academy Sciences, 102, 3691–3696.
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell, 136, 669–687.
Adai, A., Johnson, C., & Mlotshwa, S. (2005). Computational prediction of miRNAs in Arabidopsis thaliana. Genome Research, 15, 78–91.
Zhang, B. H., Pan, X. P., & Anderson, T. A. (2006). Identification of 188 conserved maize microRNAs and their targets. FEBS Letters, 580, 3753–3762.
Zhang, B. H., Wang, Q. L., Wang, K. B., Pan, X. P., Liu, F., et al. (2007). Identification of cotton microRNAs and their targets. Gene, 397, 26–37.
Yin, Z., Li, C., Han, X., & Shen, F. (2008). Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene, 414, 60–66.
Song, C., Fang, J., Li, X., Liu, H., & Chao, C. T. (2009). Identification and characterization of 27 conserved microRNAs in citrus. Planta, 230, 671–685.
Xie, F., Frazier, T. P., & Zhang, B. (2011). Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum). Gene, 473, 8–22.
Dhandapani, V., Ramchiary, N., Paul, P., Kim, J., Choi, S. H., et al. (2011). Identification of potential microRNAs and their targets in Brassica rapa L. Molecules and Cells, 32, 21–37.
Li, B., Qin, Y., Duan, H., Yin, W., & Xia, X. (2011). Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. Journal of Experimental Botany. doi:10.1093/jxb/err051.
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell, 20, 3186–3190.
Zhang, B. H., Pan, X. P., Wang, Q. L., Cobb, G. P., & Anderson, T. A. (2005). Identification and characterization of new plant microRNAs using EST analysis. Cell Research, 15, 336–360.
Sunkar, R., & Jagadeeswaran, G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology, 8, 37.
Feng, D. J., Jun, W. Y., Feng, F. X., Xia, C. J., Liang, Z., et al. (2010). Prediction of sorghum miRNAs and their targets with computational methods. Chinese Science Bulletin, 55, 1263–1270.
Lu, Y., & Yang, X. (2010). Computational identification of novel microRNAs and their targets in Vigna unguiculata. Comparative and Functional Genomics, 2010, 1–17.
Frazier, T. P., Xie, F., Freistaedter, A., Burklew, C. E., & Zhang, B. (2010). Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum). Planta, 232, 1289–1308.
Xie, F., Frazier, T. P., & Zhang, B. (2010). Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta, 232, 417–434.
Bonnet, E., Wuyts, J., Rouze, P., & Van de Peer, Y. (2004). Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics, 20, 2911–2917.
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., et al. (2002). Prediction of plant microRNA targets. Cell, 110, 513–520.
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., et al. (2005). Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 8, 517–527.
Lewis, R., Mendu, V., Mcnear, D., & Tang, G. (2010). Roles of microRNAs in plant abiotic stress. In S. M. Jain & D. S. Brar (Eds.), Molecular techniques in crop improvement (2nd ed., pp. 357–372). Cambridge, MA: Springer.
Griffiths-Jones, S. (2006). miRBase: the microRNA sequence database. Methods in Molecular Biology, 342, 129–138.
Griffiths-Jones, S., Saini, H. K., Van Dongen, S., & Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Research, 36, D154–D158.
Hofacker, I. L. (2003). Vienna RNA secondary structure server. Nucleic Acids Research, 31, 3429–3431.
Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3434.
Zuker, M., & Stiegler, P. (1981). Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Research, 9, 133–148.
Dai, X., & Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research, 39, W155–W159.
Megraw, M., Baev, V., & Rusinov, V. (2006). MicroRNA promoter element discovery in Arabidopsis. RNA, 12, 1612–1619.
Meng, Y., Shao, C., & Chen, M. (2011). Toward microRNA-mediated gene regulatory networks in plants. Brief Bioinformatics. doi:10.1093/bib/bbq091.
Zhou, L., Liu, Y., Liu, Z., Kong, D., Duan, M., et al. (2010). Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. Journal of Experimental Botany, 61, 4157–4168.
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30, 325–327.
Higo, K., Ugawa, Y., Iwamoto, M., & Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research, 27, 297–300.
Zeng, C., Wang, W., Zheng, Y., Chen, X., Bo, W., et al. (2009). Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Research, 38, 981–995.
Amiteye, S., Corral, J. M., & Sharbel, T. F. (2011). Overview of the potential of microRNAs and their target gene detection for cassava (Manihot esculenta) improvement. African Journal of Biotechnology, 11, 2562–2573.
Unver, T., Parmaksız, İ., & Dündar, E. (2010). Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Reports, 29, 757–769.
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., et al. (2008). Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 133, 116–127.
Zhang, B. H., Pan, X. P., & Stellwag, E. J. (2008). Identification of soybean microRNAs and their targets. Planta, 229, 161–182.
Okamura, K., Hagen, J. W., Duan, H., Tyler, D. M., & Lai, E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell, 130, 89–100.
Ruby, J. G., Jan, C. H., & Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature, 448, 83–86.
Moreno, M. A., Harper, L. C., Krueger, R. W., Dellaporta, S. L., & Freeling, M. (1997). Liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes and Development, 11, 616–628.
Peter, M. E. (2010). Targeting of mRNAs by multiple miRNAs: the next step. Oncogene, 29, 2161–2164.
Yin, Z., & Shen, F. (2010). Identification and characterization of conserved microRNAs and their target genes in wheat (Triticum aestivum). General Molecular Research, 9, 1186–1196.
Wu, S., Huang, S., Ding, J., Zhao, Y., Liang, L., et al. (2010). Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene, 29, 2302–2308.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., et al. (2010). MYB transcription factors in Arabidopsis. Trends in Plant Science, 15, 573–581.
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology, 10, 50.
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene, 394, 13–24.
Jofuku, K. D., Boer, B., Montagu, M. V., & Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the Homeotic Gene APETALA2. Plant Cell, 6, 1211–1225.
Mlotshwa, S., Ynag, Z., Kim, Y., & Chen, X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Molecular Biology, 61, 781–793.
Rizhsky, L., Davletova, S., Liang, H., & Mittlers, R. (2004). The zinc finger protein zat12 is required for cytosolic Ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. Journal of Biological Chemistry, 279, 11736–11743.
Achard, P., Herr, A., Baulcombe, D. C., & Harberd, N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development, 131, 3357–3365.
Acknowledgments
This research was financially supported by Mahidol University. OP was partially supported by Center of Excellence on Agricultural Biotechnology, Science and Technology Postgraduate Education and Research Development Office, Commission on Higher Education, Ministry of Education (AG-BIO/PERDO-CHE), Thailand.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2012_9521_MOESM3_ESM.pdf
Supplement Figure S1 Predicted secondary hairpin structure for 169 cassava miRNAs identified in this study. (PDF 2073 kb)
12033_2012_9521_MOESM4_ESM.tif
Supplement Figure S2 Relative transcript abundance of miR159 in the fibrous and storage root. The relative expression corresponds to the ratio of the transcript abundance of miR159 gene/transcript abundance of the small RNA U6 control gene. Data are means of three independent experiments and SE (n = 9). Different letters indicate values that are significantly different between the transcripts of the fibrous and storage root cDNA samples (Tukey’s test, one-way ANOVA; P < 0.05). (TIFF 677 kb)
Rights and permissions
About this article
Cite this article
Patanun, O., Lertpanyasampatha, M., Sojikul, P. et al. Computational Identification of MicroRNAs and Their Targets in Cassava (Manihot esculenta Crantz.). Mol Biotechnol 53, 257–269 (2013). https://doi.org/10.1007/s12033-012-9521-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9521-z