Abstract
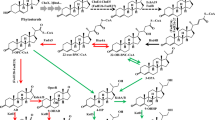
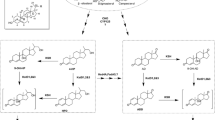
Rhodococcus rhodochrous DSM43269 is well known for its 3-ketosteroid-9α-hydroxylases. However, the function of its 3-ketosteroid-Δ1-dehydrogenases (KSDD) remains unknown. This study compared the involvement of ksdds in the strain’s androst-4-ene-3,17-dione (AD) transformation via gene deletion. The conversion was performed using AD as substrate or directly with 9α-hydroxyandrost-4-ene-3,17-dione (9α-OH-AD). The single deletion of ksdd1 or ksdd3 did not appear to result in the accumulation of 9α-OH-AD, whereas the single mutant △ksdd2 could preserve this compound to some extent. To further compare the role of ksdds in this strain, double mutants were constructed. All ksdd2 mutants combined with ksdd1 and/or ksdd3 resulted in the accumulation of 9α-OH-AD, among which the double mutant △ksdd2,3 behaved similarly to the single mutant △ksdd2 in this process. The mutant that lacked both ksdd1 and ksdd3 was still displayed, with no effect on the degradation of 9α-OH-AD. The triple mutant △ksdd1,2,3 was then constructed and exhibited the same capability as △ksdd1,2, accumulating more 9α-OH-AD than △ksdd2,3 and △ksdd2. The transcription of KSDD1 and KSDD2 increased, whereas that of KSDD3 seemed to exhibit no change, despite the use of the inducer AD or 9α-OH-AD. Thus, only ksdd1 and ksdd2 were involved in the transformation of AD to 9α-OH-AD. ksdd2 had the main role, ksdd1 had a minor effect on 9α-OH-AD degradation, and ksdd3 did not exhibit any action in this course.






Similar content being viewed by others
References
Andryushina VA, Rodina NV, Stytsenko TS, Huy LD, Druzhinina AV, Yaderetz VV, Voishvillo NE (2011) Conversion of soybean sterols into 3,17-diketosteroids using actinobacteria Mycobacterium neoaurum, Pimelobacter simplex, and Rhodococcus erythropolis. Appl Biochem Microbiol 47:270–273
Angelova B, Mutafov S, Avramova T, Stefanova L (2005) Effect of nitrogen source in cultivation medium on the 9α-hydroxylation of pregnane steroids by resting Rhodococcus sp. Cells Biotechnol Biotechnol Equip 19:113–116
De las Heras LF, van der Geize R, Drzyzga O, Perera J, Llorens JMN (2012) Molecular characterization of three 3-ketosteroid-Δ1-dehydrogenase isoenzymes of Rhodococcus ruber strain Chol-4. J Steroid Biochem Mol Biol 132:271–281
Dijkman WP, de Gonzalo G, Mattevi A, Fraaije MW (2013) Flavoprotein oxidases: classification and applications. Appl Microbiol Biotechnol 97:5177–5188
Donova MV, Dovbnya DV, Sukhodolskaya GV, Khomutov SM, Nikolayeva VM, Kwon I, Han K (2005) Microbial conversion of sterol-containing soybean oil production waste. J Chem Technol Biotechnol 80:55–60
Donova MV, Gulevskaya SA, Dovbnya DV, Puntus IF (2005) Mycobacterium sp. mutant strain producing 9α-hydroxyandrostenedione from sitosterol. Appl Microbiol Biotechnol 67:671–678
Florin C, Kӧhler T, Grandguillot M, Plesiat P (1996) Comamonas testosteroni 3-ketosteroid-delta 4(5 alpha)-dehydrogenase: gene and protein characterization. J Bacteriol 178:3322–3330
Knol J, Bodewits K, Hessels GI, Dijkhuizen L, van der Geize R (2008) 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem J 410:339–346
Larkin MJ, Kulakov LA, Allen CCR (2005) Biodegradation and Rhodococcus—masters of catabolic versatility. Curr Opin Biotechnol 16:282–290
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177
Masuda Y, Nishimura N, Tanaka K, Takakura I, Shiozaki S (1989) The production method of 9α-hydroxyandrost-4-ene-3,17-dione. Japanese Patent 1–171496
Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD (2012) Gene cluster encoding cholate catabolism in Rhodococcus spp. J Bacteriol 194:6712–6719
Morii S, Fujii C, Miyoshi T, Iwami M, Itagaki E (1998) 3-Ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous: sequencing of the genomic DNA and hyperexpression, purification, and characterization of the recombinant enzyme. J Biochem 124:1026–1032
Petrusma M, Dijkhuizen L, van der Geize R (2009) Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl Environ Microbiol 75:5300–5307
Petrusma M, Dijkhuizen L, van der Geize R (2012) Structural features in the KshA terminal oxygenase protein that determine substrate preference of 3-ketosteroid 9α-hydroxylase enzymes. J Bacteriol 194:115–121
Petrusma M, Hessels G, Dijkhuizen L, van der Geize R (2011) Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J Bacteriol 193:3931–3940
Penfield JS, Worrall LJ, Strynadka NC, Eltis LD (2014) Substrate specificities and conformational flexibility of 3-ketosteroid 9α-hydroxylases. J Biol Chem 289:25523–25536
Rodina NV, Andryushina VA, Stytsenko TS, Turova TP, Baslerov RV, Panteleeva AN, Voishvillo NE (2009) The introduction of the 9a-hydroxy group into androst-4-en-3,17-dione using a new actinobacterium strain. Appl Biochem Microbiol 45:395–400
Van der Geize R, Hessels GI, Dijkhuizen L (2002) Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid-Δ1-dehydrogenase isoenzyme. Microbiology 148:3285–3292
Van der Geize R, Hessels GI, Nienhuis-Kuiper M, Dijkhuizen L (2008) Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9α-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB. Appl Environ Microbiol 74:7197–7203
Van der Geize R, Hessels GI, van Gerwen R, Vrijbloed JW, van der Meijden P, Dijkhuizen L (2000) Targeted disruption of the kstD gene encoding a 3-ketosteroid-Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl Environ Microbiol 66:2029–2036
Yao K, Xu LQ, Wang FQ, Wei DZ (2013) Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng 15:75–87
Acknowledgments
This work was supported by the National High Technology Research and Development of China (2011AA02A211), National Natural Science Foundation of China (21276196, 21406167), Key Project of Chinese Ministry of Education (213004A), and Tianjin Programs for Science and Technology Development (15ZCZDSY00510).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Shen, Y., Qiao, Y. et al. The effect of 3-ketosteroid-Δ1-dehydrogenase isoenzymes on the transformation of AD to 9α-OH-AD by Rhodococcus rhodochrous DSM43269. J Ind Microbiol Biotechnol 43, 1303–1311 (2016). https://doi.org/10.1007/s10295-016-1804-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1804-0