Abstract
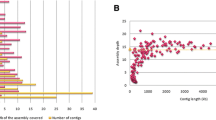
Bread wheat (Triticum aestivum L.) is one of the most important crops globally and a high priority for genetic improvement, but its large and complex genome has been seen as intractable to whole genome sequencing. Isolation of individual wheat chromosome arms has facilitated large-scale sequence analyses. However, so far there is no such survey of sequences from the A genome of wheat. Greater understanding of an A chromosome could facilitate wheat improvement and future sequencing of the entire genome. We have constructed BAC library from the long arm of T. aestivum chromosome 1A (1AL) and obtained BAC end sequences from 7,470 clones encompassing the arm. We obtained 13,445 (89.99%) useful sequences with a cumulative length of 7.57 Mb, representing 1.43% of 1AL and about 0.14% of the entire A genome. The GC content of the sequences was 44.7%, and 90% of the chromosome was estimated to comprise repeat sequences, while just over 1% encoded expressed genes. From the sequence data, we identified a large number of sites suitable for development of molecular markers (362 SSR and 6,948 ISBP) which will have utility for mapping this chromosome and for marker assisted breeding. From 44 putative ISBP markers tested 23 (52.3%) were found to be useful. The BAC end sequence data also enabled the identification of genes and syntenic blocks specific to chromosome 1AL, suggesting regions of particular functional interest and targets for future research.



Similar content being viewed by others
References
Bartoš J, Paux E, Kofler R, Havránková M, Kopecký D, Suchánková P, Šafář J, Šimková H, Town CD, Lelley T, Feuillet C, Doležel J (2008) A first survey of the rye (Secale cereale) genome composition through BAC end sequencing of the short arm of chromosome 1R. BMC Plant Biol 8:95
Berkman PJ, Skarshewski A, Lorenc MT, Lai K, Duran C, Ling EY, Stiller J, Smits L, Imelfort M, Manoli S, McKenzie M, Kubaláková M, Šimková H, Batley J, Fleury D, Doležel J, Edwards D (2011) Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotechnol J. doi:10.1111/j.1467-7652.2010.00587.x
Budak H, Shearman RC, Gulsen O, Dweikat I (2005) Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor and Appl Genetics 111:1545–1552
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinforma 10:421
Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, Couloux A, Budak H, Breen J, Pumphrey M, Liu S, Kong X, Jia J, Gut M, Brunel D, Anderson JA, Gill BS, Appels R, Keller B, Feuillet C (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22:1686–1701
Doležel J, Kubaláková M, Paux E, Bartoš J, Feuillet C (2007) Chromosome-based genomics in cereals. Chromosome Res 15:51–66
Dong Q, Lawrence CJ, Schlueter SD, Wilkerson MD, Kurtz S, Lushbough C, Brendel V (2005) Comparative plant genomics resources at PlantGDB. Plant Physiol 139:610–618
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Feuillet C, Salse J (2009) Comparative genomics in the Triticeae. Plant Genetics and Genomics 7:451–480
Gramene Release #33 (2011) Cold Spring Harbor Laboratory and Cornell University, USA. Available at: http://www.gramene.org. Accessed 20 Jul 2011
Green P (1996) phrap/cross_match/swat. Available at: http://www.phrap.org/phredphrapconsed.html. Accessed 11 Nov 2010
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Janda J, Šafář J, Kubaláková M et al (2006) Advanced resources for plant genomics: BAC library specific for the short arm of wheat chromosome 1B. Plant J 47:977–986
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kelley JM, Field CE, Craven MB, Bocskai D, Kim UJ, Rounsley SD, Adams MD (1999) High throughput direct end sequencing of BAC clones. Nucleic Acids Res 27:1539–1546
Kofler R, Schlotterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685
Kubaláková M, Vrána J, Číhalíková J, Šimková H, Doležel J (2002) Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor Appl Genet 104(8):1362–1372
Kuchel H, Langridge P, Mosionek L, Williams K, Jefferies SP (2006) The genetic control of milling yield, dough rheology and baking quality of wheat. Theor Appl Genet 112:1487–1495
Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
Li H, Tong Y, Li B, Jing R, Lu C, Li Z (2010) Genetic analysis of tolerance to photo-oxidative stress induced by high light in winter wheat (Triticum aestivum L.). J Genet Genomics 37:399–412
Mayer KF, Taudien S, Martis M, Šimková H, Suchánková P, Gundlach H, Wicker T, Petzold A, Felder M, Steuernagel B, Scholz U, Graner A, Platzer M, Doležel J, Stein N (2009) Gene content and virtual gene order of barley chromosome 1H. Plant Physiol 151:496–505
Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K (2009) TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol 150:1135–1146
Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32:D360–D363
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, Lagudah E, Somers D, Kilian A, Alaux M, Vautrin S, Bergès H, Eversole K, Appels R, Šafář J, Šimková H, Doležel J, Bernard M, Feuillet C (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322:101–104
Paux E, Faure S, Choulet F, Roger D, Gauthier V, Martinant J-P, Sourdille P, Balfourier F, Le Paslier M-C, Chauveau A, Cakir M, Gandon B, Feuillet C (2010) Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted selection in wheat. Plant Biotechnol J 8:196–210
Peng JH, Zadeh H, Lazo GR, Gustafson JP, Chao S, Anderson OD, Qi LL, Echalier B, Gill BS, Dilbirligi M, Sandhu D, Gill KS, Greene RA, Sorrells ME, Akhunov ED, Dvorak J, Linkiewicz AM, Dubcovsky J, Hossain KG, Kalavacharla V, Kianian SF, Mahmoud AA, Miftahudin CEJ, Anderson JA, Pathan MS, Nguyen HT, McGuire PE, Qualset CO, Lapitan NL (2004) Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homoeology with rice and Arabidopsis. Genetics 168:609–623
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G, Salse J (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9:473–484
Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MA, Snape JW, Angus WJ (2009) Raising yield potential in wheat. J Exp Bot 60:1899–1918
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Šafář J, Šimková H, Kubaláková M, Číhaliková J, Suchánková P, Bartoš J, Doležel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Shultz JL, Kazi S, Bashir R, Afzal JA, Lightfoot DA (2007) The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor Appl Genet 114:1081–1090
Šimková H, Šafář J, Kubaláková M, Suchánková P, Číhalíková J, Robert-Quatre H, Azhaguvel P, Weng Y, Peng J, Lapitan NLV, Ma Y, You FM, Luo M-Ch, Bartoš J, Doležel J (2011) BAC Libraries from wheat chromosome 7D: efficient tool for positional cloning of aphid resistance genes. J Biomed Biotechnol 302543
Šimková H, Číhalíková J, Vrána J, Lysák MA, Doležel J (2003) Preparation of HMW DNA from plant nuclei and chromosomes isolated from root tips. Biol Plant 46:369–373
Smith DB, Flavell RB (1975) Characterisation of wheat genome by renaturation kinetics. Chromosoma 50:223–242
The Food and Agriculture Organization of the United Nations (2009) FAO Production Statistics. Available at: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567. Accessed 11 Mar 2011
Vrána J, Kubaláková M, Šimková H et al (2000) Flow-sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156:2033–2041
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Zhang L-Y, Liu D-C, Guo X-L, Yang W-L, Sun J-Z, Wang D-W, Zhang A (2010) Genomic distribution of quantitative trait loci for yield and yield-related traits in common wheat. J Integr Plant Biol 52:996–1007
Acknowledgments
We are grateful to Prof. B. S. Gill (Kansas State University, Manhattan, USA) for seeds of the double ditelosomic line 1A of wheat T. aestivum L. cv. Chinese Spring. We thank our colleagues, Dr. Jarmila Číhalíková, Dr. Marie Kubaláková and Romana Šperková, Bc. for chromosome sorting and Jana Dostálová, Bc., Radka Tušková, Helena Tvardíková, and Dr. Marie Seifertová for excellent technical assistance in BAC library construction and Z. Weinstein for the help with the MS. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under the grant agreement no. FP7-212019.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Online resource 1
Excel spreadsheet of 362 putative SSR markers identified in chromosome 1AL BES sequences. (XLSX 42.3 kb)
Online resource 2
Excel spreadsheet of 9,338 putative ISBP markers identified in chromosome 1AL BES sequences, including details of repetitive element junctions. (XLSX 1.71 mb)
Online resource 3
Excel spreadsheet of 147 putative ISBP markers that incorporate an SSR from 1AL BES sequences, including details of repetitive element junctions and microsatellite sequences. (XLSX 42.7 kb)
Online resource 4
Excel spreadsheet of primer pairs used to test 26 ISBP, eight SSR, and ten combined ISBP/SSR markers in PCR screens, along with summary of results from amplification of both pooled BAC clones and gDNA from wheat cultivars and nullitetrasomic lines. (XLSX 14.1 kb)
Online resource 5
Table of syntenic relationships of masked 1AL BES sequences. All BES that had significant homology to coding regions in at least two sequenced grass species are shown. CDS that are out of syntenic sequence in one species are highlighted. (DOCX 21.1 kb)
Rights and permissions
About this article
Cite this article
Lucas, S.J., Šimková, H., Šafář, J. et al. Functional features of a single chromosome arm in wheat (1AL) determined from its structure. Funct Integr Genomics 12, 173–182 (2012). https://doi.org/10.1007/s10142-011-0250-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-011-0250-3