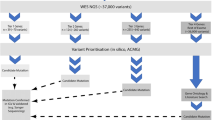
Abstract
West syndrome, which is narrowly defined as infantile spasms that occur in clusters and hypsarrhythmia on EEG, is the most common early-onset epileptic encephalopathy (EOEE). Patients with West syndrome may have clear etiologies, including perinatal events, infections, gross chromosomal abnormalities, or cases followed by other EOEEs. However, the genetic etiology of most cases of West syndrome remains unexplained. DNA from 18 patients with unexplained West syndrome was subjected to microarray-based comparative genomic hybridization (array CGH), followed by trio-based whole-exome sequencing in 14 unsolved families. We identified candidate pathogenic variants in 50 % of the patients (n = 9/18). The array CGH revealed candidate pathogenic copy number variations in four cases (22 %, 4/18), including an Xq28 duplication, a 16p11.2 deletion, a 16p13.1 deletion and a 19p13.2 deletion disrupting CACNA1A. Whole-exome sequencing identified candidate mutations in known epilepsy genes in five cases (36 %, 5/14). Three candidate de novo mutations were identified in three cases, with two mutations occurring in two new candidate genes (NR2F1 and CACNA2D1) (21 %, 3/14). Hemizygous candidate mutations in ALG13 and BRWD3 were identified in the other two cases (14 %, 2/14). Evaluating a panel of 67 known EOEE genes failed to identify significant mutations. Despite the heterogeneity of unexplained West syndrome, the combination of array CGH and whole-exome sequencing is an effective means of evaluating the genetic background in unexplained West syndrome. We provide additional evidence for NR2F1 as a causative gene and for CACNA2D1 and BRWD3 as candidate genes for West syndrome.

Similar content being viewed by others
References
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi:10.1038/nmeth0410-248
Al-Kateb H, Shimony JS, Vineyard M et al (2013) NR2F1 haploinsufficiency is associated with optic atrophy, dysmorphism and global developmental delay. Am J Med Genet 161A:377–381. doi:10.1002/ajmg.a.35650
Armentano M, Chou S-J, Tomassy GS et al (2007) COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci 10:1277–1286. doi:10.1038/nn1958
Auvin S, Holder-Espinasse M, Lamblin M-D, Andrieux J (2009) Array-CGH detection of a de novo 0.7-Mb deletion in 19p13.13 including CACNA1A associated with mental retardation and epilepsy with infantile spasms. Epilepsia 50:2501–2503. doi:10.1111/j.1528-1167.2009.02189.x
Balasubramanian M, Smith K, Mordekar SR, Parker MJ (2011) Clinical report: AN INTERSTITIAL deletion of 16p13.11 detected by array CGH in a patient with infantile spasms. Eur J Med Genet 54:314–318. doi:10.1016/j.ejmg.2011.01.008
Bosch DGM, Boonstra FN, Gonzaga-Jauregui C et al (2014) NR2F1 mutations cause optic atrophy with intellectual disability. Am J Hum Genet 94:303–309. doi:10.1016/j.ajhg.2014.01.002
Cooper GM, Goode DL, Ng SB et al (2010) Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods 7:250–251. doi:10.1038/nmeth0410-250
de Kovel CGF, Trucks H, Helbig I et al (2010) Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain 133:23–32. doi:10.1093/brain/awp262
de Ligt J, Willemsen MH, Van Bon BWM et al (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367:1921–1929. doi:10.1056/NEJMoa1206524
DePristo MA, Banks E, Poplin R et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi:10.1038/ng.806
Dimassi S, Labalme A, Lesca G et al (2014) A subset of genomic alterations detected in rolandic epilepsies contains candidate or known epilepsy genes including GRIN2A and PRRT2. Epilepsia 55:370–378. doi:10.1111/epi.12502
Dolphin AC (2012) Calcium channel auxiliary α2δ and β subunits: trafficking and one step beyond. Nat Rev Neurosci 13:542–555. doi:10.1038/nrn3311
Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS et al (2013) De novo mutations in epileptic encephalopathies. Nature 501:217–221. doi:10.1038/nature12439
Field M, Tarpey PS, Smith R et al (2007) Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. Am J Hum Genet 81:367–374. doi:10.1086/520677
Hannes FD, Sharp AJ, Mefford HC et al (2009) Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet 46:223–232. doi:10.1136/jmg.2007.055202
Heinzen EL, Radtke RA, Urban TJ et al (2010) Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet 86:707–718. doi:10.1016/j.ajhg.2010.03.018
Huang W, Luo S, Ou J et al (2014) Correlation between FMR1 expression and clinical phenotype in discordant dichorionic-diamniotic monozygotic twin sisters with the fragile X mutation. J Med Genet 51:159–164. doi:10.1136/jmedgenet-2013-101978
Jouvenceau A, Eunson LH, Spauschus A et al (2001) Human epilepsy associated with dysfunction of the brain P/Q-type calcium channel. Lancet 358:801–807. doi:10.1016/S0140-6736(01)05971-2
Kalscheuer VM, Tao J, Donnelly A et al (2003) Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet 72:1401–1411. doi:10.1086/375538
Kodera H, Nakamura K, Osaka H et al (2013) De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum Mutat 34:1708–1714. doi:10.1002/humu.22446
Kubota T, Nonoyama S, Tonoki H et al (1999) A new assay for the analysis of X-chromosome inactivation based on methylation-specific PCR. Hum Genet 104:49–55
Kumar RA, KaraMohamed S, Sudi J et al (2008) Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17:628–638. doi:10.1093/hmg/ddm376
Labrum RW, Rajakulendran S, Graves TD et al (2009) Large scale calcium channel gene rearrangements in episodic ataxia and hemiplegic migraine: implications for diagnostic testing. J Med Genet 46:786–791. doi:10.1136/jmg.2009.067967
Lux AL, Osborne JP (2004) A proposal for case definitions and outcome measures in studies of infantile spasms and West syndrome: consensus statement of the West Delphi group. Epilepsia 45:1416–1428. doi:10.1111/j.0013-9580.2004.02404.x
MacDonald JR, Ziman R, Yuen RKC et al (2014) The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42:D986–D992. doi:10.1093/nar/gkt958
McCarthy SE, Makarov V, Kirov G et al (2009) Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 41:1223–1227. doi:10.1038/ng.474
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi:10.1101/gr.107524.110
Mefford HC, Yendle SC, Hsu C et al (2011) Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol 70:974–985. doi:10.1002/ana.22645
Michaud JL, Lachance M, Hamdan FF et al (2014) The genetic landscape of infantile spasms. Hum Mol Genet. doi:10.1093/hmg/ddu199
Nagamani SCS, Erez A, Probst FJ et al (2012) Small genomic rearrangements involving FMR1 support the importance of its gene dosage for normal neurocognitive function. Neurogenetics 13:333–339. doi:10.1007/s10048-012-0340-y
Need AC, Ge D, Weale ME et al (2009) A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet 5:e1000373. doi:10.1371/journal.pgen.1000373
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814
Ng BG, Buckingham KJ, Raymond K et al (2013) Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am J Hum Genet 92:632–636. doi:10.1016/j.ajhg.2013.03.012
Ophoff RA, Terwindt GM, Vergouwe MN et al (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87:543–552
Paciorkowski AR, Thio LL, Rosenfeld JA et al (2011) Copy number variants and infantile spasms: evidence for abnormalities in ventral forebrain development and pathways of synaptic function. Eur J Hum Genet 19:1238–1245. doi:10.1038/ejhg.2011.121
Pellock JM, Hrachovy R, Shinnar S et al (2010) Infantile spasms: a U.S. consensus report. Epilepsia 51:2175–2189. doi:10.1111/j.1528-1167.2010.02657.x
Riant F, Mourtada R, Saugier-Veber P, Tournier-Lasserve E (2008) Large CACNA1A deletion in a family with episodic ataxia type 2. Arch Neurol 65:817–820. doi:10.1001/archneur.65.6.817
Rio M, Malan V, Boissel S et al (2010) Familial interstitial Xq27.3q28 duplication encompassing the FMR1 gene but not the MECP2 gene causes a new syndromic mental retardation condition. Eur J Hum Genet 18:285–290. doi:10.1038/ejhg.2009.159
Saitsu H, Kato M, Mizuguchi T et al (2008) De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet 40:782–788. doi:10.1038/ng.150
Shinawi M, Liu P, Kang S-HL et al (2010) Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 47:332–341. doi:10.1136/jmg.2009.073015
Strømme P, Mangelsdorf ME, Shaw MA et al (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445. doi:10.1038/ng862
Tang K, Xie X, Park J-I et al (2010) COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development 137:725–734. doi:10.1242/dev.040568
Timal S, Hoischen A, Lehle L et al (2012) Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum Mol Genet 21:4151–4161. doi:10.1093/hmg/dds123
Tiwari VN, Sundaram SK, Chugani HT, Huq AHMM (2013) Infantile spasms are associated with abnormal copy number variations. J Child Neurol 28:1191–1196. doi:10.1177/0883073812453496
Trevathan E, Murphy CC, Yeargin-Allsopp M (1999) The descriptive epidemiology of infantile spasms among Atlanta children. Epilepsia 40:748–751
Ullmann R, Turner G, Kirchhoff M et al (2007) Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 28:674–682. doi:10.1002/humu.20546
Vengoechea J, Parikh AS, Zhang S, Tassone F (2012) De novo microduplication of the FMR1 gene in a patient with developmental delay, epilepsy and hyperactivity. Eur J Hum Genet 20:1197–1200. doi:10.1038/ejhg.2012.78
Vergult S, Dheedene A, Meurs A et al (2014) Genomic aberrations of the CACNA2D1 gene in three patients with epilepsy and intellectual disability. Eur J Hum Genet. doi:10.1038/ejhg.2014.141
Viggiano E, Picillo E, Cirillo A, Politano L (2012) Comparison of X-chromosome inactivation in Duchenne muscle/myocardium-manifesting carriers, non-manifesting carriers and related daughters. Clin Genet. doi:10.1111/cge.12048
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi:10.1093/nar/gkq603
Weiss LA, Shen Y, Korn JM et al (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358:667–675. doi:10.1056/NEJMoa075974
Yamatogi Y, Ohtahara S (2002) Early-infantile epileptic encephalopathy with suppression-bursts, Ohtahara syndrome; its overview referring to our 16 cases. Brain Dev 24:13–23
Zhuchenko O, Bailey J, Bonnen P et al (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 15:62–69. doi:10.1038/ng0197-62
Acknowledgments
The authors thank the patients, families, and doctors who participated in this study. We thank Yoko Chiba and Kumi Ito, Miyuki Tsuda, Mami Kikuchi, Makiko Nakagawa, and Kiyotaka Kuroda for technical assistance. We also acknowledge the support of the Biomedical Research Core of Tohoku University Graduate School of Medicine. This work was supported by grant from the Ministry of Health labor and Welfare, Japan (to SK) and JSPS KAKENHI Grant Number 25461536.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all participants or their guardians.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hino-Fukuyo, N., Kikuchi, A., Arai-Ichinoi, N. et al. Genomic analysis identifies candidate pathogenic variants in 9 of 18 patients with unexplained West syndrome. Hum Genet 134, 649–658 (2015). https://doi.org/10.1007/s00439-015-1553-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-015-1553-6