Abstract
Main conclusion
The characterization and compare expression profiling of the miRNA transcriptome lay a solid foundation for unraveling the complex miRNA-mediated regulatory network in tomato resistance mechanisms against LB.
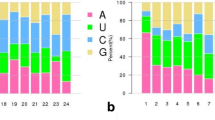
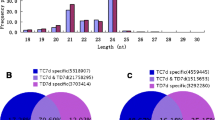
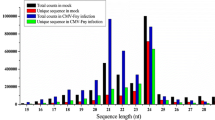
MicroRNAs (miRNAs) are a class of small endogenous non-coding RNAs with 20–24 nt. They have been identified in many plants with their diverse regulatory roles in biotic stresses. The knowledge, that miRNAs regulate late blight (LB), caused by Phytophthora infestans, is rather limited. In this study, we used miRNA-Seq to investigate the miRNA expression difference between the tomatoes treated with and without P. infestans. A total of 42,714,516 raw reads were generated from two small RNA libraries by high-throughput sequencing. Finally, 207 known miRNAs and 67 new miRNAs were obtained. The differential expression profile of miRNAs in tomato was further analyzed with twofold change (P value ≤0.01). A total of 70 miRNAs were manifested to change significantly in samples treated with P. infestans, including 50 down-regulated miRNAs and 20 up-regulated miRNAs. Moreover, a total of 73 target genes were acquired for 28 differentially expressed miRNAs by psRNATarget analysis. By enrichment pathway analysis of target genes, plant–pathogen interaction was the most highly relevant pathway which played an important role in disease defense. In addition, 30 miRNAs were selected for qRT-PCR to validate their expression patterns. The expression patterns for targets of miR6027, miR5300, miR476b, miR159a, miR164a and miRn13 were selectively examined, and the results showed that there was a negative correlation on the expression patterns between miRNAs and their targets. The targets have previously been reported to be related with plant immune and involved in plant–pathogen interaction pathway in this study, suggesting these miRNAs might act as regulators in process of tomato resistance against P. infestans. These discoveries will provide us useful information to explain tomato resistance mechanisms against LB.






Similar content being viewed by others
Abbreviations
- BGI:
-
Beijing Genomics Institute
- CMV:
-
Cucumber mosaic virus
- Cs:
-
Control samples
- ETI:
-
Effector-triggered immunity
- GO:
-
Gene Ontology
- HR:
-
Hypersensitive response
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LB:
-
Late blight
- MFE:
-
Minimum free energy
- MiRNA:
-
MicroRNA
- qRT-PCR:
-
Reverse transcription-quantitative polymerase chain reaction
- Rpi gene:
-
Resistance genes against P. infestans
- rRNA:
-
Ribosomal RNA
- snoRNA:
-
Small nuclear RNA
- snRNA:
-
Small nuclear ribonucleic acids
- TMV:
-
Tobacco mosaic virus
- ToMV:
-
Tomato mosaic virus
- tRNA:
-
Transfer RNA
- Ts:
-
Treated samples
- TuMV:
-
Turnip mosaic virus
References
Balmer D, Mauch-Mani B (2013) Small yet mighty—MicroRNAs in plant-microbe interactions. Microrna 2:72–79
Black LL, Wang TC, Hanson PM, Chen JT (1996) Late blight resistance in four wild tomato accessions: effectiveness in diverse locations and inheritance of resistance. Phytopathology 86:S24
Boccara M, Sarazin A, Thiébeauld O, Jay F, Voinnet O et al (2014) The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 10:e1003883
Bonde R, Murphy EF (1952) Resistance of certain tomato varieties and crosses to late blight. Maine Agric Exp Stn Bull 497:5–15
Chen J, Feng J, Liao Q, Chen S, Zhang J et al (2012) Analysis of tomato microRNAs expression profile induced by cucumovirus and tobamovirus infections. J Nanosci Nanotechnol 12:143–150
Cui J, Liu S, Zhang B, Wang H, Sun H et al (2014a) Transciptome analysis of the gill and swimbladder of Takifugu rubripes by RNA-Seq. PLoS One 9:e85505
Cui J, Luan Y, Wang W, Zhai J (2014b) Prediction and validation of potential pathogenic microRNAs involved in Phytophthora infestans infection. Mol Biol Rep 41:1879–1889
Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100:8024–8029
Du P, Wu J, Zhang J, Zhao S, Zheng H et al (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7:e1002176
Feng J, Liu S, Wang M, Lang Q, Jin C (2014) Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 240:1335–1352
Foolad MR, Merk HL, Ashrafi H (2008) Genetics, genomics and breeding of late blight and early blight resistance in tomato. Crit Rev Plant Sci 27:75–107
Fry WE, Goodwin SB, Matuszak JM, Spoelman LJ, Milgroom MG et al (1992) Population genetics and intercontinental migration of Phytophthora infestans. Annu Rev Phytopathol 30:107–129
Giacomelli JI, Weigel D, Chan RL, Manavella PA (2012) Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 195:766–773
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32:D109–D111
He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582:2445–2452
Huang X, Wang A, Xu X, Li J, Li N (2010) Construction of genetic linkage map and QTL analysis of Phytophthora infestans resistant gene Ph-2 in tomato. Acta Hortic Sin 37:1085–1092
Janga SC, Vallabhaneni S (2011) MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol 722:59–74
Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kim MJ, Mutschler MA (2005) Transfer to processing tomato and characterization of late blight resistance derived from Solanum pimpinellifolium L. L3708. J Am Soc Hortic Sci 130:877–884
Kim MJ, Federer W, Mutschler MA (2005) Using half-normal probability plot and regression analysis to differentiate complex traits: differentiating disease response of multigenic resistance and susceptibility in tomatoes to multiple pathogen isolates. Theor Appl Genet 112:21–29
Li J, Luan Y (2014) Molecular cloning and characterization of a pathogen-induced WRKY transcription factor gene from late blight resistant tomato varieties Solanum pimpinellifolium L3708. Physiol Mol Plant 87:25–31
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM et al (2012a) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109:1790–1795
Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q et al (2012b) Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res 40:4298–4305
Li J, Luan Y, Yin Y (2014) SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene 547:145–151
Luan Y, Wang W, Liu P (2014) Identification and functional analysis of novel and conserved microRNAs in tomato. Mol Biol Rep 41:5385–5394
Mackey D, Holt BF 3rd, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108:743–754
Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM (2013) Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci 14:7302–7326
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Merk HL, Ashrafi H, Foolad MR (2012) Selective genotyping to identify late blight resistance genes in an accession of the tomato wild species Solanum pimpinellifolium. Euphytica 187:63–75
Michaelson MJ, Price HJ, Ellison JR, Johnston JS (1991) Comparison of plant DNA contents determined by Feulgen microspectrophotometry and laser flow cytometry. Am J Bot 78:183–188
Miranda BEC, Suassuna ND, Reis A (2010) Mating type, mefenoxam sensitivity, and pathotype diversity in Phytophthora infestans isolates from tomato in Brazil. Pesq Agropec Bras 45:671–679
Park Y, Hwang J, Kim K, Kang J, Kim B (2013) Development of the gene-based SCARs for the Ph-3 locus, which confers late blight resistance in tomato. Sci Hortic 164:9–16
Pashkovskiy PP, Ryazansky SS (2013) Biogenesis, evolution, and functions of plant microRNAs. Biochem Mosc 78:627–637
Rodewald J, Trognitz B (2013) Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol Plant Pathol 14:740–757
Serrano M, Hubert DA, Dangl JL, Schulze-Lefert P, Kombrink E (2010) A chemical screen for suppressors of the avrRpm1-RPM1-dependent hypersensitive cell death response in Arabidopsis thaliana. Planta 231:1013–1023
Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA et al (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874
Socquet-Juglard D, Kamber T, Pothier JF, Christen D, Gessler C, Duffy B, Patocchi A (2013) Comparative RNA-seq analysis of early-infected peach leaves by the invasive phytopathogen Xanthomonas arboricola pv. pruni. PLoS One 8:e54196
Sun G, Luan Y, Cui J (2014) Mining and characterization of miRNAs closely associated with the pathogenicity in tomato. Yi Chuan 36:69–76
Tagami Y, Inaba N, Kutsuna N, Kurihara Y, Watanabe Y (2007) Specific enrichment of miRNAs in Arabidopsis thaliana infected with tobacco mosaic virus. DNA Res 14:227–233
Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:1
Verma SS, Rahman MH, Deyholos MK, Basu U, Kav NN (2014) Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS One 9:e86648
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Xu D, Mou G, Wang K, Zhou G (2014) MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res 190C:60–68
Yan J, Zhang H, Zheng Y, Ding Y (2014) Comparative expression profiling of miRNAs between the cytoplasmic male sterile line Meixiang A and its maintainer line Meixiang B during rice anther development. Planta 241:109–123
Yang X, Li L (2011) miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plant. Bioinformatics 27:2614–2615
Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD et al (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25:2540–2553
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z et al (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75:93–105
Zhang C, Liu L, Wang X, Vossen J, Li G et al (2014) The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor Appl Genet 127:1353–1364
Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D et al (2013) MiR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS One 8:e84390
Zuo J, Wang Y, Liu H, Ma Y, Ju Z et al (2011) MicroRNAs in tomato plants. Sci China Life Sci 54:599–605
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31272167, 31471880 and 61472061). We thank Prof. Weixing Shan from Northwest A&F, University of China for providing the pathogen materials.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luan, Y., Cui, J., Zhai, J. et al. High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans . Planta 241, 1405–1416 (2015). https://doi.org/10.1007/s00425-015-2267-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2267-7