Abstract
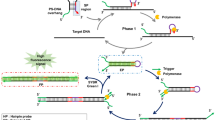
There are various ways that priming can occur in nucleic acid amplification reactions. While most reactions rely on a primer to initiate amplification, a mechanism for DNA amplification has been developed in which hairpin sequences at the 3’ terminus of a single-stranded oligonucleotide fold on themselves to initiate priming. Unfortunately, this method is less useful for diagnostic applications because the self-folding efficiency is low and only works over a narrow range of reaction temperatures. In order to adapt this strategy for analytical applications we have developed a variant that we term phosphorothioated-terminal hairpin formation and self-priming extension (PS-THSP). In PS-THSP a phosphorothioate (PS) modification is incorporated into the DNA backbone, leading to a reduction in the thermal stability of dsDNA and increased self-folding of terminal hairpins. By optimizing the number of PS linkages that are included in the initial template, we greatly increased self-folding efficiency and the range of reaction temperatures, ultimately achieving a detection limit of 1 pM. This improved method was readily adapted to the detection of single nucleotide polymorphisms and to the detection of non-nucleic acid analytes, such as alkaline phosphatase, which was quantitatively detected at a limit of 0.05 mU/mL, approximately 10-fold better than commercial assays.

Efficient self-folding by phosphorothioate (PS) modification






Similar content being viewed by others
References
Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(1):263–73.
Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–4. doi:10.1126/science.2999980.
Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–2. doi:10.1038/350091a0.
Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 2005;51:1973–81. doi:10.1373/clinchem.2005.053694.
Mukai H, Uemori T, Takeda O, et al. Highly efficient isothermal DNA amplification system using three elements of 5’-DNA-RNA-3’ chimeric primers, RNaseH and strand-displacing DNA polymerase. J Biochem (Tokyo). 2007;142:273–81. doi:10.1093/jb/mvm138.
Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification–an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–6.
Walker GT, Little MC, Nadeau JG, Shank DD. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci. 1992;89:392–6.
Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi:10.1038/sj.embor.7400200.
Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4, e204. doi:10.1371/journal.pbio.0040204.
Jung C, Chung JW, Kim UO, Kim MH, Park HG. Isothermal target and signaling probe amplification method, based on a combination of an isothermal chain amplification technique and a fluorescence resonance energy transfer cycling probe technology. Anal Chem. 2010;82:5937–43. doi:10.1021/ac100606m.
Fire A, Xu SQ. Rolling replication of short DNA circles. Proc Natl Acad Sci. 1995;92:4641–5.
Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–32. doi:10.1038/898.
Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28, e63. doi:10.1093/nar/28.12.e63.
Mitani Y, Lezhava A, Kawai Y, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–62. doi:10.1038/nmeth1007.
Kato T, Liang X, Asanuma H. Model of elongation of short DNA sequence by thermophilic DNA polymerase under isothermal conditions. Biochemistry (Mosc). 2012;51:7846–53. doi:10.1021/bi3010413.
Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–9.
Chen S, Ge B. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 2010;10:41. doi:10.1186/1471-2180-10-41.
Boczkowska M, Guga P, Stec WJ. Stereodefined phosphorothioate analogues of DNA: relative thermodynamic stability of the model PS-DNA/DNA and PS-DNA/RNA complexes. Biochemistry (Mosc). 2002;41:12483–7. doi:10.1021/bi026225z.
LaPlanche LA, James TL, Powell C, et al. Phosphorothioate-modified oligodeoxyribonucleotides. III. NMR and UV spectroscopic studies of the Rp-Rp, Sp-Sp, and Rp-Sp duplexes, [d(GGSAATTCC)]2, derived from diastereomeric O-ethyl phosphorothioates. Nucleic Acids Res. 1986;14:9081–93.
Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi:10.1146/annurev.bi.54.070185.002055.
Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents–is the bullet really magical? Science. 1993;261:1004–12. doi:10.1126/science.8351515.
Jose D, Datta K, Johnson NP, von Hippel PH. Spectroscopic studies of position-specific DNA “breathing” fluctuations at replication forks and primer-template junctions. Proc Natl Acad Sci. 2009;106:4231–6. doi:10.1073/pnas.0900803106.
Bruse SE, Moreau MP, Azaro MA, Zimmerman R, Hoffman A, Brzustowicz LM. Improvements to bead based oligonucleotide ligation SNP genotyping assays. BioTechniques. 2008;45:559–71. doi:10.2144/000112960.
Conze T, Shetye A, Tanaka Y, et al. Analysis of genes, transcripts, and proteins via DNA ligation. Annu Rev Anal Chem (Palo Alto, Calif). 2009;2:215–39. doi:10.1146/annurev-anchem-060908-155239.
Hardenbol P, Banér J, Jain M, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–8. doi:10.1038/nbt821.
Qi X, Bakht S, Devos KM, Gale MD, Osbourn A. L-RCA (ligation-rolling circle amplification): a general method for genotyping of single nucleotide polymorphisms (SNPs). Nucleic Acids Res. 2001;29, e116.
Shin GW, Chung B, Jung GY, Jung GY. Multiplex ligase-based genotyping methods combined with CE. Electrophoresis. 2014;35:1004–16. doi:10.1002/elps.201300361.
Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci. 1991;88:189–93.
Barany F. The ligase chain reaction in a PCR world. Genome Res. 1991;1:5–16. doi:10.1101/gr.1.1.5.
Zhang S, Wu Z, Shen G, Yu R. A label-free strategy for SNP detection with high fidelity and sensitivity based on ligation-rolling circle amplification and intercalating of methylene blue. Biosens Bioelectron. 2009;24:3201–7. doi:10.1016/j.bios.2009.03.012.
Acknowledgments
This work was supported by the National Institutes of Health [5 R01 AI092839, 5 R01 GM094933]. We sincerely thank Caitlin Sanford for her editing services.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Published in the topical collection Isothermal Nucleic Acid Amplification in Bioanalysis with guest editor Maria Jesus Lobo Castañón.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 591 kb)
Rights and permissions
About this article
Cite this article
Jung, C., Ellington, A.D. A primerless molecular diagnostic: phosphorothioated-terminal hairpin formation and self-priming extension (PS-THSP). Anal Bioanal Chem 408, 8583–8591 (2016). https://doi.org/10.1007/s00216-016-9479-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9479-y