Abstract
The assessment of nucleic acid degradation levels is essential to guarantee the accuracy and reproducibility of the experimental results. This step becomes more important when working with fixed and paraffin-embedded (FPE) tissues, where the variable and unpredictable effects of the fixation processing on macromolecule preservation are dependent not only on the intrinsic properties of the fixative but also on the tissue-related preanalytical variables. In this chapter, several methods for the assessment of nucleic acid integrity are described, the focus being especially on their accuracy in FPE and suitability according to the availability and nature of the starting material. In particular, some PCR-based assays have also been reported as useful methods for integrity estimation when DNA/RNA starting material is very poor. Moreover, special attention has been given to RNA quality assessment, which commonly relies on methods based on the detection of ribosomal RNA as an indirect measure of target mRNA integrity. In this context, the limitations of these approaches in the quality assessment of RNA extracted from FPE have been pointed out, suggesting that PCR-based strategies, based on the detection of specific mRNA targets, still remain the best methods for the analysis of RNA isolated from archival material.
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction and Purpose
The level of degradation of nucleic acids, and especially of RNA, is a very important parameter to assess assay amenability when using fixed and paraffin-embedded (FPE) samples. The reason for this is that it’s not a very well predictable phenomenon and its variable effects may false the results of qualitative and quantitative analysis on DNA and RNA. This variability depends on both biological and technical factors (type of tissue, fixation parameters, sample processing...). The degradation levels can be measured by different approaches and they are chosen according to the nature of the nucleic acid.
2 Agarose Gel Electrophoresis for DNA
Indicative integrity of genomic DNA can be checked on a 0.4% agarose gel with ethidium bromide and the level of degradation can be graded according to the electrophoretic migration of sample DNA in comparison to a known molecular weight marker (usually λDNA/HindIII fragments). It can be run with or without previous enzymatic digestion depending on the expected level of degradation and the consequent length of DNA fragments present in the sample. In the first case, the degradation rate can be defined by comparing the band pattern between an intact DNA and the unknown sample; in the second case, the index of degradation is determined by the presence of a smearing pattern. For more details on the gel preparation, see also Appendix A.
3 Denaturing Agarose Gel Electrophoresis for RNA
The quality of mRNA from fresh samples (cultured cell lines, fresh frozen tissue samples…) can be easily assessed by running total RNA in a denaturing gel stained with ethidium bromide. This method relies on the assumption that rRNA quality and quantity reflect those of the underlying mRNA population. RNA integrity evaluation is based on the intensity ratio between the 28S and 18S bands (2:1 ratio is considered an index for intact RNA) and on the observation of the rRNA degradation pattern. Anyway, it is not clear how rRNA degradation actually reflects the quality of the underlying mRNA population, since in certain conditions rRNA can turn over more rapidly than mRNA [1]. Besides, visual assessment of the 28S:18S rRNA ratio on agarose gels is not an objective parameter because the appearance of rRNA bands is affected by the electrophoretic conditions, the amount of loaded RNA, and the saturation of ethidium bromide fluorescence.
This approach is not suitable when RNA integrity assessment is performed on archival tissues because large amounts of input RNA are needed and the ribosomal bands are rarely detectable due to the high levels of RNA degradation.
3.1 Reagents
All reagents should be RNase freeFootnote 1 and DEPC treated
-
DEPC H 2 O
-
10% SDS or alternatively 1 M NaOH or H 2 O 2
-
Agarose powder electrophoresis grade
-
37% Formaldehyde
-
10X MOPS buffer: 0.2 M MOPS, 0.05 M sodium acetate pH 6, 0.01 M EDTA
-
RNA loading buffer. Footnote 2 For 1.5 ml: 720 μl formamide, 160 μl 10X MOPS buffer, 260 μl 37% formaldehyde, 80 μl 1% blue bromophenol, 100 μl 80% glycerol, 180 μl DEPC H2O
-
Ethidium Bromide, 10 mg/ml (stock solution). Footnote 3 Stock solution of EtBr should be stored at 4°C in a dark bottle
-
Running buffer: 1/10 of 37% formaldehyde in volume, 1X MOPS
3.2 Equipment
-
Disinfected adjustable pipettes,Footnote 4 range: 2–20 μl, 20–200 μl, 100–1,000 μl
-
Nuclease-free aerosol-resistant pipette tips
-
0.5 or 1.5 ml tubes (autoclaved)
-
Centrifuge suitable for centrifugation/spinning at 13,200 or 14,000 rpm
-
Thermoblock
-
Electrophoretic apparatus with spacers
-
Power supply
-
UV transilluminator
3.3 Method
-
Wash the electrophoretic apparatus and spacers with 1% SDS (alternatively it is possible to use 1 M NaOH or 7% H2O2), cover and leave for 2 h under a fume hood. Wash with distilled H2O, followed by sterile H2O and finally DEPC H2O.
-
Prepare the 1% agarose gel by melting 0.5 g agarose in 42.3 ml DEPC H2O. When cooled, add 5 ml 10X MOPS and 2.7 ml 37% formaldehyde. Pour the solution in the apparatus.Footnote 5
-
To prepare the RNA samples, use 5–50 μg total RNA. Mix 2–10 μl of RNA solution with 5 μl of RNA loading buffer. Adjust the volume with DEPC H2O to a final volume of 20 μl.
-
Denature RNA for 5′ at 65°C and chill on ice.
-
Add 1 μl ethidium bromide previously diluted to 200 μg/ml and load the samples into the gel.
-
Separate the products at 70 V in 1X running buffer until the dye reaches three-fourths of the gel length.
-
Check the gel under the UV light.Footnote 6 Two main bands corresponding to 28S and 18S rRNA should be detected. Acquire the intensity of the bands by a computer-based system. A 2:1 ratio between the two bands is considered an index of intact RNA. The presence of a smear and/or the absence of the ribosomal bands is an index of RNA degradation (Fig. 17.1).
Fig. 17.1
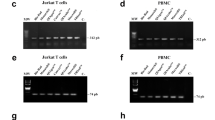
4 PCR Amplification of DNA Fragments of Increasing Length
DNA quality from archival samples can be assessed by PCR amplification selecting fragments of increasing length. One commercial method that can be used for this purpose is the “Specimen Control Size Ladder” that is provided by InVivo Scribe Technologies. The kit consists of a master mix composed of a buffered magnesium solution, deoxynucleotides, and multiple primer pairs that allow multiplex amplification of five DNA fragments ranging from 100 to 600 base pairs. With the “Specimen Control Size Ladder” – Unlabeled both a conventional agarose and acrylamide gel can be run to visualize the results (Fig. 17.2).
5 RT-PCR Amplification of mRNA Fragments of Increasing Length
The setting up of this method is very easy. Messenger RNA integrity is normally evaluated by RT-PCR amplification of a housekeeping gene selecting a ladder of mRNA fragments of increasing length. Reverse transcription is normally performed using MMLV enzyme combined with the random-hexamer priming strategy. The specific primer pairs used in the amplification step are designed for fragments of progressive length, preferably keeping either primer constant (Fig. 17.3). The length of the amplicons can be chosen arbitrarily, but is usually between 200 and 3,000 bp for DNA, and between 60 and 1,000 bp for RNA [2]. The major advantage of mRNA integrity determination is that the results are independent from rRNA integrity. This method can be useful in FFPE samples as it requires small amounts of starting material. (For the setting of this experiment follow the rules reported in Chaps. 19 and 25).
6 3′/5′ mRNA Integrity Assay
This method is specific to assess mRNA degradation levels and represents a variation of the previous approach (“RT-PCR amplification of mRNA fragments of increasing length”). It measures the integrity of the selected target gene (usually a highly expressed housekeeping gene such as GAPDH and beta-actin), which is expected to reflect the integrity of the entire mRNA subpopulation in the sample [3, 4]. In this assay reverse transcription is performed with oligo-dT and Superscript II/III. A real-time PCR reaction is carried out for three 60–80 bp-long amplicons along the transcript using three differently labeled TaqMan probes. Usually, amplicons are located at 100, 400–500, and 800–1,000 bases from the 3′ end of the gene, corresponding to the so-called 3′, intermediate and 5′ fragment, respectively. The quantitative ratios between the Ct values of the 3′ and 5′ amplicons reflect the degradation level and the efficiency of the RT enzyme to proceed along the transcript. The higher the RNA fragmentation, the lower the RT efficiency, and consequently, the PCR yield. A 3′:5′ ratio of about 1 indicates high RNA integrity, whereas progressive ratios indicate increasing degradation (Fig. 17.4). For details regarding the setting of the TaqMan-based experiments and interpretation of the results, see Chap. 25, Sect. 25.2.
Quantitative ratios calculated from the Ct values associated with the 3′ fragment and the two fragments located at increasing length from the 3′ end of the gene. Ratios of RNA isolated from fresh frozen sample usually correspond to 1 and progressively increase in RNA extracted from formalin-fixed samples (in this example cultured cell samples were progressively formalin fixed from 1 to 24 h)
7 Agilent Bioanalyzer 2100
See http://www.chem.agilent.com/Scripts/PDS.asp?lPage=51 for more details
This microfluidic capillary electrophoresis system by Agilent Technologies is based on fluorescent dyes binding to nucleic acid and allows the checking of not only DNA and RNA integrity, but also nucleic acid quantity (as already reported in Chap. 16, Sect. 16.2.2.5). When RNA integrity is assessed, three types of data analysis can be performed by this instrument. The first is the visual analysis of the electropherogram (“visual method”). It assigns a categorization of the RNA degradation levels (e.g., from 1 to 5) [5], taking into account the ribosomal peak height, the baseline flatness, and the presence of additional peaks between the ribosomal peaks (Fig. 17.5). Alternatively, two automatically calculated parameters can be used. The first one is the 28S/18S ratio (“Ratio method”). A ribosomal band ratio of (28S:18S) between 0.7 and 2.5 is considered to be typical of good-quality RNA. The second one is the RIN (RNA Integrity Number). It takes into account the whole electropherogram, including in the analysis the total RNA ratio (the fraction of the area in the region of 18S and 28S compared to the total area under the curve), the fast region (the area between 18S peak and the marker), and the marker height.Footnote 7 The RIN range is between 1 and 10 (RIN 1 corresponds to completely degraded RNA, whereas a RIN 10 is achieved for an intact RNA sample). Values equal or higher than 6 are associated with good-quality RNA [6]. The Agilent Bioanalyzer system offers several advantages. It requires a very small amount of starting material, allowing assaying DNA and RNA quality even in small samples. Furthermore, interpretation of integrity is standardized and for this reason it is comparable among different laboratories. However, this approach appears more suitable for fresh frozen samples than for archival specimens because of the high degradation levels of nucleic acids isolated from fixed and embedded material. When RNA integrity is assessed in these specimens, 28S/18S ratios are usually around 0 and RIN are usually between 1 and 3.5 [1]. These low values, however, do not preclude the use of nucleic acids isolated from archival samples in PCR-based assays. For details about sample preparation and loading, follow the manufacturer’s instructions.
Electropherograms representing different levels of RNA degradation. (a) Nearly intact RNA (isolated from fresh frozen tissue) (b) Highly degraded RNA (isolated from FFPE tissue). These traces can be used to evaluate RNA integrity by visual assessment (operator-dependent), 28S/18S ratio and RIN number (automatically calculated by the software)
Notes
- 1.
Sterile, disposable plasticware should be preferably used because it is RNase free. If general laboratory glassware or plasticware is used, it should be presoaked in 0.1% DEPC-treated H2O for 2 h at 37°C. The DEPC treated-items should be rinsed thoroughly with DEPC H2O and then autoclaved. Alternatively, glassware can be put in an oven at 250°C for at least 2 h.
- 2.
It can be stored at 4°C for a maximum of 2 weeks or at –20°C for a maximum of 2 months.
- 3.
EtBr is a potentially carcinogenic compound. Always wear gloves. Used EtBr solutions must be collected in containers for chemical waste and discharged according to the local hazardous chemical disposal procedures.
- 4.
Clean the pipettes with a disinfectant (e.g. Meliseptol®rapid) and leave them under the UV lamp for at least 10 min. Alternatively, it is possible to autoclave the pipette depending on the provider instructions.
- 5.
Formaldehyde is carcinogenic; for this reason, gels should be prepared and run under a fume hood.
- 6.
If the gel must be submitted to further analysis (e.g., Northern blotting), deposit it on a clean plastic wrap before checking on the transilluminator.
- 7.
The marker is an internal standard that is used to align the ladder analysis with the individual sample analysis. The RNA ladder is a mixture of six RNA transcripts of well-defined size and total concentration that is run first from the ladder well. Both marker and ladder are provided in the kit.
References
Dotti I, Bonin S, Basili G, Nardon E, Balani A, Siracusano S, Zanconati F, Palmisano S, De Manzini N, Stanta G (2010) Effects of formalin, methacarn and finefix fixatives on RNA preservation. Diagn Mol Pathol 19(2):112–122
Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive’s tissues. Diagn Mol Pathol. 2006 Jun;15(2):115–23
Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–82
Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, Woosley JT, Thomas NE, Perou CM, Sandler RS, Sharpless NE. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007 Apr;87(4):383–91
Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C (2005) Towards standardization of quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33(6):e56
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T (2006) The RIN: an integrity number for assigning integrity values to measurements. BMC Mol Biol 7:3
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Dotti, I., Bonin, S. (2011). Integrity Assessment of Nucleic Acids. In: Stanta, G. (eds) Guidelines for Molecular Analysis in Archive Tissues. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-17890-0_17
Download citation
DOI: https://doi.org/10.1007/978-3-642-17890-0_17
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-17889-4
Online ISBN: 978-3-642-17890-0
eBook Packages: MedicineMedicine (R0)