Abstract
In this work we present extensions for Radial Basis Function networks to improve their ability for discrete and continuous pain intensity estimation. Besides proposing a mid-level fusion scheme, the use of standardization and unconventional loss functions are covered. We show that RBF networks can be improved in this way and present extensive experimental validation to support our findings on a multi-modal dataset.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Radial Basis Function
- Mahalanobis Distance
- Radial Basis Function Neural Network
- Radial Basis Function Network
- Skin Conductance Level
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Physiological and pathophysiological pains are survival mechanisms generated by the brain in order to stimulate protective behavior. Accordingly, pain can be considered as a measure of medical health and elementary pain based treatments have been shown to be beneficial in 20 % to 70 % of the cases [15]. However, not all clinical patients are able to identify their level of pain such as neonates and somnolent patients. It is expressed that 30 % to 70 % of the patients were suffering from pain after undergoing surgery [22]. Therefore, automatic pain estimation as an element of electronic health surveillance has recently received increasing attention.
Initial pain quantification methods mostly used facial expressions for pain estimation. Different feature sets such as Principal Components and Gabor features had been combined with Support Vector Machines (SVMs) and Relevance Vector Machine (RVM) classifiers in a binary classification scenario (pain versus no pain) in [2, 5, 12, 14]. Three class classification and continuous quantification of pain had also been done in [9, 13], respectively.
Multi-modal pain classification [21] in binary and multi-class scenarios illustrates that promising cues exist in bio-physiological signals as well as video to recognize pain. Improved results accompanied by continuous pain estimation verified the merit of the bio-physiological signals in [7, 8]. The recent study in [7] confirmed that pain is a very personal sensation and some subjects’ physiological behavior has been shown to be more similar to specific subjects than to others.
The single modality approaches relying only on the facial expressions are highly sensitive to face detection. In the clinical setup a permanent face detection is rarely feasible. Thus, uninterrupted pain estimation is not always possible using video features. Furthermore, in the case of pain estimation using dynamic classification approaches (such as Echo State Networks [6]), miss detections not only reduce the estimation accuracy of the current sample but also introduce uncertainty to the memory of the subsequent samples. It should be noted that commercially available fitness watches can measure some of the physiological signals such as blood volume pressure and galvanic skin response. Pain evaluators using bio-physiology can simply be attached to such devices in order to improve health monitoring.
The Radial Basis Function (RBF) neural networks [18] are used in this paper for continuous and discrete pain estimation through the physiological signals. The multi-class problem is investigated alongside the binary classification of the pain versus no pain. Back-propagation, Huber loss function, personalization and various fusion schemes are introduced and combined with the RBF networks to improve the accuracy of the pain estimation. Additionally, the application of the RBF nueral network is extended form classification to regression and the pain is estimated continuously.
The remainder of this paper is organized as follows: The BioVid experiment and database on which the experiments here are based is explained in Sect. 2, the physiological signal segmentation and feature extraction is presented in Sect. 3. We briefly explain Radial Basis Networks (RBF) in Sect. 4, before the experimental results are provided in Sect. 5 and the paper is concluded in Sect. 6.
2 The BioVid Experiment and Pain Database
The BioVid heat experiment [21] was conducted in 2013 at the University of Ulm. The main idea of the experiment was to stimulate the subjects with different heat levels to record bio-physiological signals namely, electrocardiography (ECG), electromyography (EMG) and skin conductance level (SCL) signals as well as video. One of the experiment objectives is to predict the pain level by multimodal processing of the physiological signals and the video. Furthermore, the stimulus signal is also recorded during the experiment.
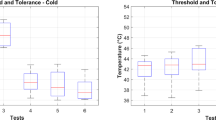
Heat was applied to 96 subjects of this experiment using a thermode which was attached to their right hand (Fig. 1a). Subsequently, different levels of heat were considered as various levels of pain. In order to reduce the different tolerances of subjects, a calibration had been done before the experiment. Initially the temperature of 32 \(^\circ \)C is assumed as the pain free temperature or level 0 of pain. Then the temperature is increased gradually and the subject is asked to react once he/she feels the pain. The initial feeling threshold of heat is considered as the first level of pain. The temperature is raised again afterwards until the highest endurable level is declared by the subject. The highest temperature is assumed as pain level 4. Two other levels of stimulation were selected corresponding to the temperatures linearly spaced between level 1 and 4.
After the calibration procedure the subjects were stimulated with different temperatures evaluated as various pain levels. The selection of heat levels was random but the subjects were stimulated 20 times with each heat level in the first part of the experiment. The duration of the stimulation was 4 s and there was a 8–12 s stimulation free time between each two consecutive stimulus (Fig. 1b). The same experiment had been done in another section after 40 min of heat relaxation. The facial electromyography (EMG) signals were recorded in the second part in addition to the previous modalities of the first part of the experiment. Complementary details of the BioVid experiment are provided in [21].
(a) The thermode attached to the right wrist (the image is taken from [21]). (b) The heat stimulation baseline temperature (\(T_0\)) versus the pain threshold (\(T_1\)) and maximum endurance level (\(T_{4}\)). The signal segmentation for feature extraction is illustrated by the green window. Image taken from [7].
3 Bio-physiological Feature Extraction
After the experiment, by removing all the defected signals, a dataset with 86 participants was left for analysis. Moreover, misplacement of the biopotential sensors as well as prolongation of the experiment degrades the pain estimation performance in the second part. Therefore, we have just analyzed the physiological signals of the first part of pain stimulation.
Considering the delay in the response of the sympathetic nervous system, the biopotential signals were segmented by a 1 s latency relative to the beginning of the stimulus (Fig. 1b) [7]. According to the physiological basis and motivations a window of length 5.5 s was used for the signal segmentation. It should be mentioned that the signals for the level zero of pain (baseline) were extracted within duration of 5.5 s right before the first stimulus pulse after the pain level one. This assumption was considered to minimize the effect of previous stimulations on the baseline.
A wide range of features were extracted from the segmented signals in order to be used in the classification and regression tasks. These features can be categorized into four main mathematical groups as follows:
-
The time domain features such as Willison amplitude, V-order, log detector and so on aimed at measuring the intensity [16].
-
The frequency group features include mode, mean, central and median frequencies, bandwidth and low pass to very low pass ratio in order to evaluate the rate of the vibration of muscles [10].
-
The stationary features are the stationary mean, variance, median and area targeted at measuring the stability of the statistical properties of signals [20].
-
The entropy features are Shannon, sample, approximation, fuzzy and spectral entropies [4] and the Shannon entropy of peak frequency shifting [3].
In addition to the mathematically based features, the physiological signals can also be processed according to their psychophysiological characteristic. For instance, the Blood Volume Pulse (BVP) signals can be analyzed in terms of R-R intervals or QRS complexes. Similarly, the skin conductance signals can be decomposed into phasic and tonic components based on physiological motivations [1]. The final sets of features used in this paper per modality are as follows:
-
EMG: The set of the EMG features includes the statistical, time, frequency and stationarity related features of the electromyography signals [8].
-
BVP1: The approximation coefficients of a four level wavelet decomposition using Daubechies wavelets of the blood volume pulse signals [23].
-
BVP2: This feature set includes the amplitude of different points, time differences and angles of the PQRST complex of the heart signals [19].
-
SCL1: The same features as for the EMG channel and 7 additional statistical features including skewness and kurtosis of the SCL signals formed this set of the features.
-
SCL2: The last set of the features are based on the phasic and tonic decomposition of the SCL signals. These features include the number of SCL responses, latency of the first response, average of the phasic driver and etc. The mentioned features are derived according to the Ledalab project [1].
4 Radial Basis Function (RBF) Neural Networks
The remainder of this paper focuses on discrete and continuous quantification of pain using Radial Basis Function (RBF) neural networks. Besides classification, we extend the application of RBF networks to regression problems.
As shown in Fig. 2 the RBF networks are a three layer neural network including input, hidden (Radial Basis Function) and output layers. The general learning procedure of this network can consist of three different steps:
-
The first step is an unsupervised learning procedure to learn the Radial Basis Function centers and width which is mainly done using the k-means algorithm.
-
The second step is a supervised learning procedure to learn the output weights to create an efficient mapping from activation values to the target vector of the classes. This step is supposed to be done using pseudo-inversion.
-
The third step is the simultaneous optimization of all parameters including output weights, Radial Basis Function center and width using the backpropagation algorithm.
In previous literatures (for instance [18]) the 1-phase and 2-phase learning procedures were also considered. However, it is notable that 1-phase learning using backpropagation and proper initialization of the RBF parameters is possible. Moreover, the RBF network can be trained using k-means and the pseudo-inverse in a 2-phase learning procedure.
Before explaining the algorithms, we would like to express the notation that will be used later on in this report which is corresponding to Fig. 2 as follows:
-
The \(\mu ^{th}\) input feature vector is denoted by \(\varvec{x}^\mu \in \mathrm I\!R^p\) with \(\mu = 1,\ldots ,M\).
-
The \(j^{th}\) cluster center of length p is expressed by \(\varvec{c}_j\in \mathrm I\!R^p\) with overall K clusters.
-
The \(j^{th}\) Radial Basis Function corresponding to an arbitrary modality (EMG, BVP or SCL) is denoted by \(\varphi _j^{modality}\) and \(\zeta _0 = 1\) is the bias term.
-
The output of the RBF neural network and the target vector are denoted by \(F_k(\varvec{x})\) and \(Y_k(\varvec{x})\) respectively. Here, we consider L output classes.
4.1 The Gaussian Kernel, Width and Distance
After the k-means algorithm which calculates the cluster centers and members, we need to define a function to evaluate the distance of a data point \(\varvec{x}^\mu \) from a cluster center \(\varvec{c}_j\). Let’s define a positive definite matrix \(\varvec{R}_j\) and accordingly assume the distance of a data feature vector and cluster center as:
The different situations for the matrix \(\varvec{R}_j\) which lead to the use of various distances, are as follows:
-
1.
The most general form of \(\varvec{R}_j\) is the inverse of the covariance matrix of the data samples within a cluster.
-
2.
Using the main diagonal of the inverse of covariance matrix leads to the Mahalanobis distance.
-
3.
Instead of \(\varvec{R}_j\), a factor of identical matrix \(\varvec{I}\) can be used for each cluster.
-
4.
Alternatively, a global scaling parameter can be used for all clusters.
It is notable that the inverse of the covariance matrices of the feature vectors are not always positive definite. In the case of a non-positive definite covariance matrix Eq. 1 can not be used directly to determine the distance. Finally the Gaussian function for each cluster center is defined as:
The output of the Gaussian function is called the activation value and denoted by \(\varvec{h}_j\) in Eq. 2. The output weights of the Radial Basis Function neural networks are trained using the pseudo-inverse matrix as a supervised learning based on the target labels.
4.2 Early, Mid-level and Late Fusions
According to the multi-modality of the BioVid database and the experiment, various fusion schemes can be applied. The first scheme is the early fusion where the new feature vectors are formed by concatenation of all the features of all modalities for every sample and the classifier is trained based on these new feature vectors.
The other commonly used method is late fusion where different classifiers are trained for each modality and the decisions of all classifiers are fused for the final decision. In the case of late fusion we used the mean of the confidence values of all classifiers as a criterion for the final decision in this paper. It should be considered that before late fusion, in order to avoid random extreme values, a soft-max function is applied to the output of each RBF classifiers.
In addition to the commonly used fusion methods, we propose mid-level fusion for RBF neural networks in this paper. In this case the unsupervised part of the learning procedure (clustering) has been done in every feature set independently. Afterwards, the activation values from all clusters of different modalities ([\(\varphi _1^{EMG},\ldots ,\varphi _K^{EMG},\varphi _1^{BVP},\ldots ,\varphi _K^{BVP},\varphi _1^{SCL},\ldots ,\varphi _K^{SCL}\)]) are concatenated and finally the output layer weights are trained based on the activations of all clusters of every feature set.
4.3 Using Huber Loss Function in RBF Neural Networks
The normal Radial Basis Function (RBF) neural networks perform inefficiently in the case of non-standardized data. After analyzing the activation values, it turns out that for some data points just one feature can dominate the whole activation value in a cluster. In other words, if one data sample is close to a cluster center in all feature dimensions expect one but very far just in that one dimension, the activation value of that cluster will be dominated by that single feature. To compensate for this effect and to obtain a robust classifier we used the Huber loss function instead of the euclidean distance in the argument of the Gaussian function. The Huber loss function is depicted in Fig. 3 for different free parameters (\(\delta \)) and can be mathematically expressed as follows:
4.4 Backpropagation
Backpropagation can be used to improve the training procedure. The main idea of the approach is to minimize an error function which is the sum of squared deviations of the output of the classes (\(\varvec{F}_k\)) from the target vectors (\(\varvec{y}_k\)). This error function is defined as [18]:
The minimization task for Eq. 4 has been done iteratively using the gradient descent procedure. In other words, all the parameters of the network are shifted in the direction in which the error function has the quickest decrease. The value of this shift is computed as a multiplication of the learning rate (\(\eta \)) to the partial derivative of the error function with respect to the parameter that is optimized. The adaption rules for different parameters of the RBF network are as follows [18]:
where, \(\sigma _{ij}\) is the width of the Gaussian kernel of the \(j^{th}\) cluster on the \(i^{th}\) coordinate of the features. In fact the \(\sigma _{ij}\) are the elements on the main diagonal of the matrix \(\varvec{R}_j\) mentioned in Eq. 1.
4.5 Regression
The regression task is aimed at evaluating an input feature vector in terms of a continuous regression value instead of discrete classes. Accordingly, we might expect only one output neuron in the output layer of Fig. 2. Consequently we will have only one target vector in the regression task which should include all the classes. We define the target vector of the regression task (\(\varvec{y}_{reg}\)) as:
Based on the regression target vector defined in Eq. 8 the output weights will be trained. Ultimately, the output of the regression task could be scaled and mapped to the desired values which are temperatures.
5 Simulations Results
The classification and regression tasks in this paper are defined as leave one subject out cross validation. In other words, in both cases the samples related to one participant are left out of the training set for testing and then the neural network or classifier is trained using the rest of the dataset. The left out subject is used as the test case for the trained classifier or regressor. All the present results in the following classification tables are the average value of such a cross validation classification for all 86 participants of the experiment.
5.1 Baseline Results
First, as a baseline to compare the RBF classifier results, we conduct the classification using RBF classifier with 50 clusters on the extracted features. The result of the classification for some pairs of the classes and the multi-class classifier are illustrated in Table 1. The baseline result shows that the classification accuracy barely surpasses 80 % in the binary problem of the pain level 0 versus 4 and the five class classification only shows 32.1 % of accuracy. It is notable that the skin conductance level features derived from phasic and tonic decomposition is the best feature set in all the classification scenarios and the result of early fusion hardy exceed the a-priori probabilities of the classification tasks.
After exploring the baseline result one can simply realize that the skin conductance level features are much more discriminant for this classification tasks than the other channels. However, it is interesting to notice that the accuracy of the RBF network considerably deteriorates for the case of non-standard or badly scaled features. That is to say, applying the same kernel width to high dimensional feature vectors in which the variances of the features are considerably different will not lead to an accurate RBF classifier. For instance, we can observe such an effect in the SCL1 feature set which includes a wide range of features with different means and variances. This effect can be observed in the poor early fusion results of the RBF neural networks as well. However, in the case of the other SCL2 feature set which is not badly scaled the result in much more accurate. In the reminder of this paper we will propose standardization schemes to improve the classification accuracy.
5.2 Standardization and Personalization
The Radial Basis Function neural networks are sensitive to non-standardized data. Therefore, we will standardize the features in two ways before conducting the classification.
Standardization. Initially, we will standardize the data in a way that every final feature has a mean of zero and variance of one over the whole data set for all participants. This method is the normal standardization. The classification results are provided in Table 2. As can be seen in the Table 2, standardization improves the maximum of the classification accuracy in all tasks. It should be mentioned that standardization raises the classification accuracy in the case of early fusion noticeably.
Personalization. Another level of standardization called person dependent standardization or personalization can be conducted to improve the classification results. In this proposed method every feature for each independent person is standardized to zero mean and unit variance. The result of the classification using personalized features for RBF neural networks classifier is presented in Table 3.
The classification result using standardized and personalized feature clarifies that both methods lead to improvement in the pain estimation accuracy. Whereas, the personalized features perform better than the standardized ones. The reason is the difference in baseline of the physiological signals for the different participants. For example, a participant with higher regular heart rate might have a heart rate corresponding to what the others experience in the first level of pain. The mid-level fusion structure proposed in this project shows a promising accuracy is some cases such as classification of the class 0 versus 4 based on personalized data. Furthermore, the late fusion in two binary classification scenarios and early fusion for multi-class problem outperforms the classification only based on the skin conductance level features.
5.3 Further Optimizations
Up to this point we have some acceptable results using RBF classifiers for standardized and personalized features. However, classification of non-standardized features is still an open problem.
Mahalanobis Distance. According to the literature a possible solution for dealing with non-standard or badly scaled features is using the Mahalanobis distance. The classification results using Mahalanobis distance is shown in Table 4. Furthermore, the classification results for non-standardized data using Mahalanobis distance is provided in Table 5.
Using Mahalanobis distance increases the classification accuracy in the multi-class and class 0 versus 4 tasks up to 37.3 % and 84.9 % respectively. Moreover, RBF neural networks with this scale-invariant distance outperform (Table 5) the same network with euclidean distance (Table 1) on non-standardized data. The maximum classification accuracy based on the non-standardized features of all tasks improves using Mahalanobis distance. The most notable improvement can be seen in the classification result of early fusion scheme and the first feature set for skin conductance level in Table 5 compared with Table 1.
It will also be informative to investigate at least one of the confusion matrices of the these classifiers. The confusion matrix of the multi-class classification task using Mahalanobis distance is provided in Table 6. It is obvious that the RBF classifier doesn’t produce all the classes as an output with the same probability. The reason is that the middle levels of pain including level 1 to 3 are highly overlapping. Additionally, Table 6 shows that there is a high probability of confusion between the pain threshold (level 1) and no pain. The best classified levels on pain in the multi-class problem are highest level of pain (level 4) and the no pain (class 0). Learning the RBF neural network based on fusion mapping can improve this situation [17].
Huber Loss Function. The free parameter of the Huber loss function can be changed to reach a good classification result. Figure 4 illustrates the classification result of non-standardized data using the RBF neural network for different free parameters (\(\delta \)). It can be observed from Fig. 4 that using the RBF neural networks with the free parameter of \(\delta = 3\) can improve the maximum classification accuracy for the non-standardized. The classification result using Huber loss function in RBF network is represented in Table 7. It is visible that the classification accuracy of the pain level 0 versus 4 is improved up to 1.8 % compared to the accuracy achieved by using Mahalanobis distance for non-standardized features. However, the classification accuracy for the multi-class problem is slightly below what have been reached through using Mahalanobis distance.
Backpropagation. The performance of the Radial Basis Function neural networks can be improved using back-propagation. The considerable drawback of this method is the high computational complexity. Some classification results for RBF using back-propagation for standardized data are illustrated in Table 8. It is observable in Table 8 that personalization accompanied by Mahalanobis distance and backpropagation boosts the baseline result of classification of class 0 versus 4 from 80.1 % up to 85 %.
It can be seen from Table 8 that the backpropagation optimization improves the classification accuracy of the RBF classifier compared with the base one (Table 4) in most of the cases. The base network here is a Radial Basis Function (RBF) neural network with Mahalanobis distance. It is beneficial to use various learning rates for different parameters (cluster center position, cluster width and output weights) optimized by the backpropagation as well.
5.4 Regression
Finally, we deal with the continuous estimation of the pain level using regression and RBF neural networks. For this purpose, we primarily used the initial setup of the RBF neural network with personalization. As we mentioned regarding signal segmentations, the training sample signals are segmented starting 1 s after the stimulus starts. Accordingly, a suitable shifting in labels is required.
There are two reasons for such a shift. First, as mentioned above the regressor is not trained based on the stimulus sequence but the segments with a 1 s lag relative to the start of the stimulus. In other words, the regression scenario here is not a sequence mapping from feature space to the stimulus which can compensate the lag of the predictions. Secondly, the heat stimulation does not affect all the physiological signal immediately and even with the same delay in time. Complementary research can be done to optimize the window length and delay of the signal segments compared to the stimulus for feature extraction.
The result of the regression for continuous pain level estimation versus the stimulus signal with and without proper shifting are illustrated and compared in Fig. 5.
Ultimately, different regression results are quantitatively compared in term of root mean squared error, normal and concordance correlation coefficients [11]. The result of this comparison is also provided in Table 9. The temporal miss-match between the prediction and label is obvious from the negative correlation coefficient reported in the Table 9. Having said that, after a suitable temporal shift the predictions shows a promising correlation of 0.48 with the labels. However, the concordance correlation coefficient can be improved by using more robust scaling methods.
6 Conclusion
The continuous and discrete pain level estimation is presented in this paper according to the BioVid heat database using Radial Basis Function neural networks. The RBF classifier using Mahalanobis distance alongside personalization reaches a good compromise between accuracy and complexity. However, using backpropagation accompanied by a softmax at the output of the classifier improves the classification performance at the expense of imposing a high amount of computational complexity. The proposed mid-level fusion outperforms the early and late fusion for some tasks. The Huber loss function improves the classification results slightly for standard features. Ultimately, it is recommended to use a combination of the aforementioned methods to run the RBF networks up to highest performance. We showed that RBF networks have potential for classifying and predicting discrete and continuous pain intensity levels. Furthermore, the network provides informative output about the samples such as confidence values. This information can be used in active learning and also used to built another layer for late fusion using pseudo inversion to improve the fusion results in multi-modal scenarios.
References
Benedek, M., Kaernbach, C.: Decomposition of skin conductance data by means of nonnegative deconvolution. Psychophysiology 47(4), 647–658 (2010)
Brahnam, S., Chuang, C.F., Shih, F.Y., Slack, M.R.: Machine recognition and representation of neonatal facial displays of acute pain. Artif. Intell. Med. 36(3), 211–222 (2006)
Cao, C., Slobounov, S.: Application of a novel measure of EEG non-stationarity as ‘Shannon-entropy of the peak frequency shifting’ for detecting residual abnormalities in concussed individuals. Clin. Neurophysiol. 122(7), 1314–1321 (2011)
Chen, W., Zhuang, J., Yu, W., Wang, Z.: Measuring complexity using FuzzyEn, ApEn, and SampEn. Med. Eng. Phys. 31(1), 61–68 (2009)
Gholami, B., Haddad, W.M., Tannenbaum, A.R.: Agitation and pain assessment using digital imaging. In: Conference Proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE Engineering in Medicine and Biology Society, Conference, vol. 2009. NIH Public Access (2009)
Jaeger, H.: The “echo state” approach to analysing and training recurrent neural networks. GMD report 148, GMD - German National Research Institute for Computer Science (2001)
Kächele, M., Thiam, P., Amirian, M., Schwenker, F., Palm, G.: Methods for person-centered continuous pain intensity assessment from bio-physiological channels. IEEE J. Sel. Top. Sign. Process. 13(9), 1–11 (2016)
Kächele, M., Thiam, P., Amirian, M., Werner, P., Walter, S., Schwenker, F., Palm, G.: Multimodal data fusion for person-independent, continuous estimation of pain intensity. In: Iliadis, L., Jayne, C. (eds.) EANN 2015. CCIS, vol. 517, pp. 275–285. Springer, Heidelberg (2015)
Kaltwang, S., Rudovic, O., Pantic, M.: Continuous pain intensity estimation from facial expressions. In: Bebis, G., et al. (eds.) ISVC 2012, Part II. LNCS, vol. 7432, pp. 368–377. Springer, Heidelberg (2012)
Kim, J., André, E.: Emotion recognition based on physiological changes in music listening. IEEE Trans. Pattern Anal. Mach. Intell. 30(12), 2067–2083 (2008)
Lin, L.I.: A concordance correlation coefficient to evaluate reproducibility. Biometrics 45(1), 255–268 (1989)
Littlewort, G.C., Bartlett, M.S., Lee, K.: Automatic coding of facial expressions displayed during posed and genuine pain. Image Vis. Comput. 27(12), 1797–1803 (2009)
Lucey, P., Cohn, J.F., Prkachin, K.M., Solomon, P.E., Chew, S., Matthews, I.: Painful monitoring: automatic pain monitoring using the UNBC-McMaster shoulder pain expression archive database. Image Vis. Comput. 30(3), 197–205 (2012)
Lucey, P., Cohn, J.F., Prkachin, K.M., Solomon, P.E., Matthews, I.: Painful data: the UNBC-McMaster shoulder pain expression archive database. In: 2011 IEEE International Conference on Automatic Face and Gesture Recognition and Workshops (FG 2011), pp. 57–64. IEEE (2011)
Moore, R.A., Wiffen, P.J., Derry, S., Maguire, T., Roy, Y.M., Tyrrell, L.: Non-prescription (OTC) oral analgesics for acute pain-an overview of cochrane reviews. Status and Date, New published in (10) (2013)
Phinyomark, A., Limsakul, C., Phukpattaranont, P.: A novel feature extraction for robust EMG pattern recognition. arXiv preprint arXiv:0912.3973 (2009)
Schwenker, F., Dietrich, C., Thiel, C., Palm, G.: Learning of decision fusion mappings for pattern recognition. Int. J. Artif. Intell. Mach. Learn. (AIML) 6, 17–21 (2006)
Schwenker, F., Kestler, H.A., Palm, G.: Three learning phases for radial-basis-function networks. Neural Netw. 14(4), 439–458 (2001)
Tang, X., Shu, L.: Classification of electrocardiogram signals with RS and quantum neural networks. Int. J. Multimedia Ubiquit. Eng. 9(2), 363–372 (2014)
Tkach, D., Huang, H., Kuiken, T.A.: Research study of stability of time-domain features for electromyographic pattern recognition. J. Neuroeng. Rehabil. 7, 21 (2010)
Walter, S., Gruss, S., Limbrecht-Ecklundt, K., Traue, H.C., Werner, P., Al-Hamadi, A., Diniz, N., da Silva, G.M., Andrade, A.O.: Automatic pain quantification using autonomic parameters. Psychol. Neurosci. 7(3), 363 (2014)
Wiebalck, A., Vandermeulen, E., Van Aken, H., Vandermeersch, E.: Ein konzept zur verbesserung der postoperativen schmerzbehandlung. Der Anaesthesist 44(12), 831–842 (1995)
Zhao, Q., Zhang, L.: ECG feature extraction and classification using wavelet transform and support vector machines. In: International Conference on Neural Networks and Brain, 2005, ICNN&B 2005, vol. 2, pp. 1089–1092. IEEE (2005)
Acknowledgement
This paper is based on work done within the Transregional Collaborative Research Centre SFB/TRR 62 Companion-Technology for Cognitive Technical Systems funded by the German Research Foundation (DFG). Markus Kächele is supported by a scholarship of the Landesgraduiertenförderung Baden-Württemberg at Ulm University. The work is furthermore supported by the SenseEmotion project funded by the German Ministry of Science, Research and Arts and was performed using the computational resource bwUniCluster funded by the Ministry of Science, Research and Arts and the Universities of the State of Baden-Württemberg, Germany, within the framework program bwHPC.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Amirian, M., Kächele, M., Schwenker, F. (2016). Using Radial Basis Function Neural Networks for Continuous and Discrete Pain Estimation from Bio-physiological Signals. In: Schwenker, F., Abbas, H., El Gayar, N., Trentin, E. (eds) Artificial Neural Networks in Pattern Recognition. ANNPR 2016. Lecture Notes in Computer Science(), vol 9896. Springer, Cham. https://doi.org/10.1007/978-3-319-46182-3_23
Download citation
DOI: https://doi.org/10.1007/978-3-319-46182-3_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46181-6
Online ISBN: 978-3-319-46182-3
eBook Packages: Computer ScienceComputer Science (R0)